Characterization of enhancer binding by the Vibrio cholerae flagellar regulatory protein FlrC
- PMID: 15838043
- PMCID: PMC1082837
- DOI: 10.1128/JB.187.9.3158-3170.2005
Characterization of enhancer binding by the Vibrio cholerae flagellar regulatory protein FlrC
Abstract
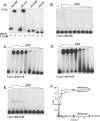
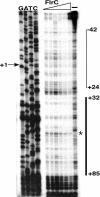
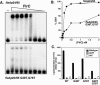
The human pathogen Vibrio cholerae is a highly motile organism by virtue of a polar flagellum, and motility has been inferred to be an important aspect of virulence. It has previously been demonstrated that the sigma(54)-dependent activator FlrC is necessary for both flagellar synthesis and for enhanced intestinal colonization. In order to characterize FlrC binding, we analyzed two FlrC-dependent promoters, the highly transcribed flaA promoter and the weakly transcribed flgK promoter, utilizing transcriptional lacZ fusions, mobility shift assays, and DNase I footprinting. Promoter fusion studies showed that the smallest fragment with wild-type transcriptional activity for flaAp was from positions -54 to +137 with respect to the start site, and from -63 to +144 for flgKp. Gel mobility shift assays indicated that FlrC binds to a fragment containing the region from positions +24 to +95 in the flaAp, and DNase I footprinting identified a protected region between positions +24 and +85. Mobility shift and DNase I footprinting indicated weak binding of FlrC to a region downstream of the flgKp transcription start site. These results demonstrate a relatively novel sigma(54)-dependent promoter architecture, with the activator FlrC binding downstream of the sigma(54)-dependent transcription start sites. When the FlrC binding site(s) in the flaA promoter was moved a large distance (285 bp) upstream of the transcription start site of either flaAp or flgKp, high levels of FlrC-dependent transcription resulted, indicating that this binding region functions as an enhancer element. In contrast, the relatively weak FlrC binding site(s) in the flgK promoter failed to function as an enhancer element at either promoter, suggesting that FlrC binding strength contributes to enhancer activity. Our results suggest that the differences in FlrC binding to various flagellar promoters results in the differences in transcription levels that mirror the relative requirement for the flagellar components within the flagellum.
Figures
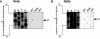



References
-
- Correa, N. E., C. M. Lauriano, R. McGee, and K. E. Klose. 2000. Phosphorylation of the flagellar regulatory protein FlrC is necessary for Vibrio cholerae motility and enhanced colonization. Mol. Microbiol. 35:743-755. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

