Expression of the melC operon in several Streptomyces strains is positively regulated by AdpA, an AraC family transcriptional regulator involved in morphological development in Streptomyces coelicolor
- PMID: 15838045
- PMCID: PMC1082821
- DOI: 10.1128/JB.187.9.3180-3187.2005
Expression of the melC operon in several Streptomyces strains is positively regulated by AdpA, an AraC family transcriptional regulator involved in morphological development in Streptomyces coelicolor
Abstract
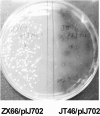
Dark brown haloes of melanin around colonies are an easily visualized phenotype displayed by many Streptomyces strains harboring plasmid pIJ702 carrying the melC operon of Streptomyces antibioticus IMRU3270. Spontaneous melanin-negative mutants of pIJ702 occur with a frequency of ca. 1%, and often mutation occurs in the melC operon, which removes the BglII site as part of an inverted repeat. Other melanin-negative mutations seem to occur spontaneously in Streptomyces lividans, resulting in white colonies from which intact, melanin-producing pIJ702 can be isolated by introduction into a new host. S. lividans ZX66 was found to be such a mutant and to have a secondary mutation influencing expression of the melC operon on the chromosome. A 3.3-kb DNA fragment was isolated from its progenitor strain, JT46, and a gene able to restore melC operon expression was found to encode a member of an AraC family of transcriptional regulators, which was equivalent to AdpA(c) in Streptomyces coelicolor and therefore was designated AdpA(l). Lack of melC operon expression was correlated with a single A-to-C transversion, which altered a single key amino acid residue from Thr to Pro. The transcription of the melC operon was found to be greatly reduced in the adpA mutant background. The counterpart gene (adpA(a)) in the S. antibioticus strain in which the melC operon carried on pIJ702 originated was also isolated and was found to have an identical regulatory role. Thus, we concluded that the melC operon is under general direct positive control by AdpA family proteins, perhaps at the transcriptional level and certainly at the translational level via bldA, in Streptomyces.
Figures



Similar articles
-
Improved vectors for transcriptional/translational signal screening in corynebacteria using the melC operon from Streptomyces glaucescens as reporter.Arch Microbiol. 2003 Jul;180(1):53-9. doi: 10.1007/s00203-003-0560-5. Epub 2003 Jun 7. Arch Microbiol. 2003. PMID: 12802479
-
Autorepression of AdpA of the AraC/XylS family, a key transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus.J Mol Biol. 2005 Jul 1;350(1):12-26. doi: 10.1016/j.jmb.2005.04.058. J Mol Biol. 2005. PMID: 15907934
-
DNA-binding specificity of AdpA, a transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus.Mol Microbiol. 2004 Jul;53(2):555-72. doi: 10.1111/j.1365-2958.2004.04153.x. Mol Microbiol. 2004. PMID: 15228534
-
Negative regulation of the heat shock response in Streptomyces.Arch Microbiol. 2001 Oct;176(4):237-42. doi: 10.1007/s002030100321. Arch Microbiol. 2001. PMID: 11685367 Review.
-
AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus.Biosci Biotechnol Biochem. 2005 Mar;69(3):431-9. doi: 10.1271/bbb.69.431. Biosci Biotechnol Biochem. 2005. PMID: 15784968 Review.
Cited by
-
Extracellular and intracellular polyphenol oxidases cause opposite effects on sensitivity of Streptomyces to phenolics: a case of double-edged sword.PLoS One. 2009 Oct 14;4(10):e7462. doi: 10.1371/journal.pone.0007462. PLoS One. 2009. PMID: 19826489 Free PMC article.
-
Unlocking Streptomyces spp. for use as sustainable industrial production platforms by morphological engineering.Appl Environ Microbiol. 2006 Aug;72(8):5283-8. doi: 10.1128/AEM.00808-06. Appl Environ Microbiol. 2006. PMID: 16885277 Free PMC article.
-
Melanin production as a visual indicator of conjugal transfer in Streptomyces.J Appl Genet. 2020 May;61(2):299-301. doi: 10.1007/s13353-020-00540-0. Epub 2020 Jan 13. J Appl Genet. 2020. PMID: 31933172
-
Microbial ultraviolet sunscreens.Nat Rev Microbiol. 2011 Oct 3;9(11):791-802. doi: 10.1038/nrmicro2649. Nat Rev Microbiol. 2011. PMID: 21963801 Review.
-
Natural pigments derived from plants and microorganisms: classification, biosynthesis, and applications.Plant Biotechnol J. 2025 Feb;23(2):592-614. doi: 10.1111/pbi.14522. Epub 2024 Dec 6. Plant Biotechnol J. 2025. PMID: 39642082 Free PMC article. Review.
References
-
- Altschul, S. F., and E. V. Koonin. 1998. Iterated profile searches with PSI-BLAST—a tool for discovery in protein databases. Trends Biochem. Sci. 23:444-447. - PubMed
-
- Bernan, V., D. Filpula, W. Herber, M. Bibb, and E. Katz. 1985. The nucleotide sequence of the tyrosinase gene from Streptomyces antibioticus and characterization of the gene product. Gene 37:101-110. - PubMed
-
- Bierman, M., R. Logan, K. O′Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

