Molecular determinants for PspA-mediated repression of the AAA transcriptional activator PspF
- PMID: 15838051
- PMCID: PMC1082823
- DOI: 10.1128/JB.187.9.3238-3248.2005
Molecular determinants for PspA-mediated repression of the AAA transcriptional activator PspF
Abstract
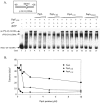
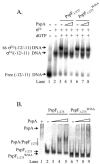
The Escherichia coli phage shock protein system (pspABCDE operon and pspG gene) is induced by numerous stresses related to the membrane integrity state. Transcription of the psp genes requires the RNA polymerase containing the sigma(54) subunit and the AAA transcriptional activator PspF. PspF belongs to an atypical class of sigma(54) AAA activators in that it lacks an N-terminal regulatory domain and is instead negatively regulated by another regulatory protein, PspA. PspA therefore represses its own expression. The PspA protein is distributed between the cytoplasm and the inner membrane fraction. In addition to its transcriptional inhibitory role, PspA assists maintenance of the proton motive force and protein export. Several lines of in vitro evidence indicate that PspA-PspF interactions inhibit the ATPase activity of PspF, resulting in the inhibition of PspF-dependent gene expression. In this study, we characterize sequences within PspA and PspF crucial for the negative effect of PspA upon PspF. Using a protein fragmentation approach, we show that the integrity of the three putative N-terminal alpha-helical domains of PspA is crucial for the role of PspA as a negative regulator of PspF. A bacterial two-hybrid system allowed us to provide clear evidence for an interaction in E. coli between PspA and PspF in vivo, which strongly suggests that PspA-directed inhibition of PspF occurs via an inhibitory complex. Finally, we identify a single PspF residue that is a binding determinant for PspA.
Figures

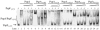




References
-
- Aseeva, E., F. Ossenbuhl, L. A. Eichacker, G. Wanner, J. Soll, and U. C. Vothknecht. 2004. Complex formation of Vipp1 depends on its alpha-helical PspA-like domain. J. Biol. Chem. 279:35535-35541. - PubMed
-
- Bates, P. A., L. A. Kelley, R. M. MacCallum, and M. J. E. Sternberg. 2001. Enhancement of protein modelling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Proteins 5(Suppl.):39-46. - PubMed
-
- Beloin, C., J. Valle, P. Latour-Lambert, P. Faure, M. Kzreminski, D. Balestrino, J. A. Haagensen, S. Molin, G. Prensier, B. Arbeille, and J.-M. Ghigo. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51:659-674. - PubMed
-
- Bogan, A. A., and K. S. Thorn. 1998. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 280:1-9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

