Multistress regulation in Escherichia coli: expression of osmB involves two independent promoters responding either to sigmaS or to the RcsCDB His-Asp phosphorelay
- PMID: 15838058
- PMCID: PMC1082829
- DOI: 10.1128/JB.187.9.3282-3286.2005
Multistress regulation in Escherichia coli: expression of osmB involves two independent promoters responding either to sigmaS or to the RcsCDB His-Asp phosphorelay
Abstract
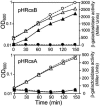
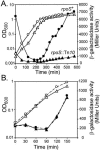
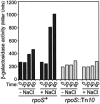
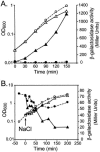
Transcription of the Escherichia coli osmB gene is induced by several stress conditions. osmB is expressed from two promoters, osmBp1 and osmBp2. The downstream promoter, osmBp2, is induced after osmotic shock or upon entry into stationary phase in a sigma(S)-dependent manner. The upstream promoter, osmBp1, is independent of sigma(S) and is activated by RcsB, the response regulator of the His-Asp phosphorelay signal transduction system RcsCDB. RcsB is responsible for the induction of osmBp1 following treatment with chlorpromazine. Activation of osmBp1 by RcsB requires a sequence upstream of its -35 element similar to the RcsB binding site consensus, suggesting a direct regulatory role. osmB appears as another example of a multistress-responsive gene whose transcription involves both a sigma(S)-dependent promoter and a second one independent of sigma(S) but controlled by stress-specific transcription factors.
Figures
References
-
- Bouvier, J., S. Gordia, G. Kampmann, R. Lange, R. Hengge-Aronis, and C. Gutierrez. 1998. Interplay between global regulators of Escherichia coli: effect of RpoS, Lrp and H-NS on transcription of the gene osmC. Mol. Microbiol. 28:971-980. - PubMed
-
- Carballes, F., C. Bertrand, J. P. Bouche, and K. Cam. 1999. Regulation of Escherichia coli cell division genes ftsA and ftsZ by the two-component system rcsC-rcsB. Mol. Microbiol. 34:442-450. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

