Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes
- PMID: 15839726
- PMCID: PMC1084335
- DOI: 10.1371/journal.pbio.0030157
Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes
Abstract
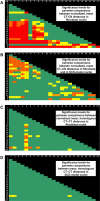
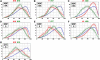
Studies of higher-order chromatin arrangements are an essential part of ongoing attempts to explore changes in epigenome structure and their functional implications during development and cell differentiation. However, the extent and cell-type-specificity of three-dimensional (3D) chromosome arrangements has remained controversial. In order to overcome technical limitations of previous studies, we have developed tools that allow the quantitative 3D positional mapping of all chromosomes simultaneously. We present unequivocal evidence for a probabilistic 3D order of prometaphase chromosomes, as well as of chromosome territories (CTs) in nuclei of quiescent (G0) and cycling (early S-phase) human diploid fibroblasts (46, XY). Radial distance measurements showed a probabilistic, highly nonrandom correlation with chromosome size: small chromosomes-independently of their gene density-were distributed significantly closer to the center of the nucleus or prometaphase rosette, while large chromosomes were located closer to the nuclear or rosette rim. This arrangement was independently confirmed in both human fibroblast and amniotic fluid cell nuclei. Notably, these cell types exhibit flat-ellipsoidal cell nuclei, in contrast to the spherical nuclei of lymphocytes and several other human cell types, for which we and others previously demonstrated gene-density-correlated radial 3D CT arrangements. Modeling of 3D CT arrangements suggests that cell-type-specific differences in radial CT arrangements are not solely due to geometrical constraints that result from nuclear shape differences. We also found gene-density-correlated arrangements of higher-order chromatin shared by all human cell types studied so far. Chromatin domains, which are gene-poor, form a layer beneath the nuclear envelope, while gene-dense chromatin is enriched in the nuclear interior. We discuss the possible functional implications of this finding.
Figures






References
-
- van Driel R, Fransz PF, Verschure PJ. The eukaryotic genome: A system regulated at different hierarchical levels. J Cell Sci. 2003;116:4067–4075. - PubMed
-
- Cremer T, Kurz A, Zirbel R, Dietzel S, Rinke B, et al. Role of chromosome territories in the functional compartmentalization of the cell nucleus. Cold Spring Harb Symp Quant Biol. 1993;58:777–792. - PubMed
-
- Cremer T, Kreth G, Koester H, Fink RH, Heintzmann R, et al. Chromosome territories, interchromatin domain compartment, and nuclear matrix: An integrated view of the functional nuclear architecture. Crit Rev Eukaryot Gene Expr. 2000;10:179–212. - PubMed
-
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. - PubMed
-
- Parada LA, Misteli T. Chromosome positioning in the interphase nucleus. Trends Cell Biol. 2002;12:425. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

