Expression, purification, and characterization of SARS coronavirus RNA polymerase
- PMID: 15840516
- PMCID: PMC7111802
- DOI: 10.1016/j.virol.2005.02.017
Expression, purification, and characterization of SARS coronavirus RNA polymerase
Abstract
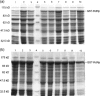
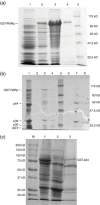
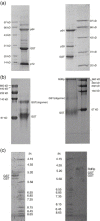
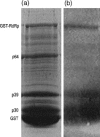
The RNA-dependent RNA polymerase (RdRp) of SARS coronavirus (SARS-CoV) is essential for viral replication and a potential target for anti-SARS drugs. We report here the cloning, expression, and purification of the N-terminal GST-fused SARS-CoV RdRp and its polymerase catalytic domain in Escherichia coli. During purification, the full-length GST-RdRp was found to cleave into three main fragments: an N-terminal p12 fragment, a middle p30 fragment, and a C-terminal p64 fragment comprising the catalytic domain, presumably due to bacterial proteases. Biochemical assays show that the full-length GST-RdRp has RdRp activity and the p64 and p12 fragments form a complex that exhibits comparable RdRp activity, whereas the GST-p64 protein has no activity, suggesting that the p12 domain is required for polymerase activity possibly via involvement in template-primer binding. Nonnucleoside HIV-1 RT inhibitors are shown to have no evident inhibitory effect on SARS-CoV RdRp activity. This work provides a basis for biochemical and structural studies of SARS-CoV RdRp and for development of anti-SARS drugs.
Figures

References
-
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for Design of anti-SARS drugs. Science. 2003;300:1763–1767. - PubMed
-
- Chan L., Reddy T.J., Proulx M., Das S.K., Pereira O., Wang W., Siddiqui A., Yannopoulos C.G., Poisson C., Turcotte N., Drouin A., Alaoui-Ismaili M.H., Bethell R., Hamel M., L'Heureux L., Bilimoria D., Nguyen-Ba N. Identification of N,N-disubstituted phenylalanines as a novel class of inhibitors of hepatitis C NS5B polymerase. J. Med. Chem. 2003;46:9489–9495. - PubMed
-
- Das K., Ding J., Hsiou Y., Clark A.D., Jr., Moereels H., Koymans L., Andries K., Pauwels R., Janssen P.A.J., Boyer P.L., Clark P., Smith R.H., Jr., Kroeger Smith M.B., Michejda C.J., Hughes S.H., Arnold E. Crystal structures of 8-Cl and 9-Cl TIBO complexed with wild-type HIV-1 RT and 8-Cl TIBO complexed with the Tyr181Cys HIV-1 RT drug-resistant mutant. J. Mol. Biol. 1996;264:1085–1100. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

