Perturbational profiling of a cell-line model of tumorigenesis by using metabolic measurements
- PMID: 15840712
- PMCID: PMC1087961
- DOI: 10.1073/pnas.0502267102
Perturbational profiling of a cell-line model of tumorigenesis by using metabolic measurements
Abstract
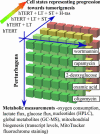
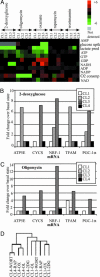
Weinberg and coworkers have used serial transduction of a human, primary fibroblast cell line with the catalytic domain of human telomerase, large T antigen, small T antigen, and an oncogenic allele of H-ras to study stages leading toward a fully transformed cancerous state. We performed a three-dimensional screening experiment using 4 cell lines, 5 small-molecule perturbagens (2-deoxyglucose, oxamate, oligomycin, rapamycin, and wortmannin), and a large number of metabolic measurements. Hierarchical clustering was performed to obtain signatures of the 4 cell lines, 24 cell states, 5 perturbagens, and a number of metabolic parameters. Analysis of these signatures and sensitivities of the cell lines to the perturbagens provided insights into the bioenergetic states of progressively transformed cell lines, the effect of oncogenes on small-molecule sensitivity, and global physiological responses to modulators of aerobic and anaerobic metabolism. We have gained insight into the relationship between two models of carcinogenesis, one (the Warburg hypothesis) based on increased energy production by glycolysis in cancer cells in response to aberrant respiration, and one based on cancer-causing genes. Rather than being opposing models, the approach described here suggests that these two models are interlinked. The cancer-causing genes used in this study appear to increase progressively the cell's dependence on glycolytic energy production and to decrease its dependence on mitochondrial energy production. However, mitochondrial biogenesis appears to have a more complex dependence, increasing to its greatest extent at an intermediate degree of transduction rather than at the fully transformed state.
Figures


References
-
- Hahn, W. C., Counter, C. M., Lundberg, A. S., Beijersbergen, R. L., Brooks, M. W. & Weinberg, R. A. (1999) Nature 400, 464-468. - PubMed
-
- Dolma, S., Lessnick, S. L., Hahn, W. C. & Stockwell, B. R. (2003) Cancer Cell 3, 285-296. - PubMed
-
- Norrbom, J., Sundberg, C. J., Ameln, H., Kraus, W. E., Jansson, E. & Gustafsson, T. (2004) J. Appl. Physiol. 96, 189-194. - PubMed
-
- Leung, L., Kwong, M., Hou, S., Lee, C. & Chan, J. Y. (2003) J. Biol. Chem. 278, 48021-48029. - PubMed
-
- Mootha, V. K., Lindgren, C. M., Eriksson, K. F., Subramanian, A., Sihag, S., Lehar, J., Puigserver, P., Carlsson, E., Ridderstrale, M., Laurila, E., et al. (2003) Nat. Genet. 34, 267-273. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

