Gain-of-function amino acid substitutions drive positive selection of FGFR2 mutations in human spermatogonia
- PMID: 15840724
- PMCID: PMC1087921
- DOI: 10.1073/pnas.0500267102
Gain-of-function amino acid substitutions drive positive selection of FGFR2 mutations in human spermatogonia
Abstract
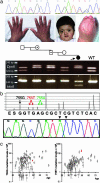
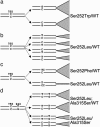
Despite the importance of mutation in genetics, there are virtually no experimental data on the occurrence of specific nucleotide substitutions in human gametes. C>G transversions at position 755 of FGF receptor 2 (FGFR2) cause Apert syndrome; this mutation, encoding the gain-of-function substitution Ser252Trp, occurs with a birth rate elevated 200- to 800-fold above background and originates exclusively from the unaffected father. We previously demonstrated high levels of both 755C>G and 755C>T FGFR2 mutations in human sperm and proposed that these particular mutations are enriched because the encoded proteins confer a selective advantage to spermatogonial cells. Here, we examine three corollaries of this hypothesis. First, we show that mutation levels at the adjacent FGFR2 nucleotides 752-754 are low, excluding any general increase in local mutation rate. Second, we present three instances of double-nucleotide changes involving 755C, expected to be extremely rare as chance events. Two of these double-nucleotide substitutions are shown, either by assessment of the pedigree or by direct analysis of sperm, to have arisen in sequential steps; the third (encoding Ser252Tyr) was predicted from structural considerations. Finally, we demonstrate that both major alternative spliceforms of FGFR2 (Fgfr2b and Fgfr2c) are expressed in rat spermatogonial stem cell lines. Taken together, these observations show that specific FGFR2 mutations attain high levels in sperm because they encode proteins with gain-of-function properties, favoring clonal expansion of mutant spermatogonial cells. Among FGFR2 mutations, those causing Apert syndrome may be especially prevalent because they enhance signaling by FGF ligands specific for each of the major expressed isoforms.
Figures



References
-
- Crow, J. F. (2000) Nat. Rev. Genet. 1, 40-47. - PubMed
-
- Li, W.-H., Yi, S. & Makova, K. (2002) Curr. Opin. Genet. Dev. 12, 650-656. - PubMed
-
- Hurst, L. D. (2003) in Nature Encyclopaedia of the Human Genome, ed. Cooper, D. N. (Nature Publishing Group, London), Vol. 4, pp. 218-222.
-
- Moloney, D. M., Slaney, S. F., Oldridge, M., Wall, S. A., Sahlin, P., Stenman, G. & Wilkie, A. O. M. (1996) Nat. Genet. 13, 48-53. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

