Structural basis for the interaction of Asf1 with histone H3 and its functional implications
- PMID: 15840725
- PMCID: PMC1087920
- DOI: 10.1073/pnas.0500149102
Structural basis for the interaction of Asf1 with histone H3 and its functional implications
Abstract
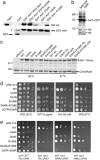
Asf1 is a conserved histone chaperone implicated in nucleosome assembly, transcriptional silencing, and the cellular response to DNA damage. We solved the NMR solution structure of the N-terminal functional domain of the human Asf1a isoform, and we identified by NMR chemical shift mapping a surface of Asf1a that binds the C-terminal helix of histone H3. This binding surface forms a highly conserved hydrophobic groove surrounded by charged residues. Mutations within this binding site decreased the affinity of Asf1a for the histone H3/H4 complex in vitro, and the same mutations in the homologous yeast protein led to transcriptional silencing defects, DNA damage sensitivity, and thermosensitive growth. We have thus obtained direct experimental evidence of the mode of binding between a histone and one of its chaperones and genetic data suggesting that this interaction is important in both the DNA damage response and transcriptional silencing.
Figures
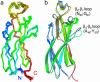



References
-
- Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. (1997) Nature 389, 251-260. - PubMed
-
- Loyola, A. & Almouzni, G. (2004) Biochim. Biophys. Acta 1677, 3-11. - PubMed
-
- Verreault, A. (2000) Genes Dev. 14, 1430-1438. - PubMed
-
- Tyler, J. K., Adams, C. R., Chen, S. R., Kobayashi, R., Kamakaka, R. T. & Kadonaga, J. T. (1999) Nature 402, 555-560. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

