Prp8 protein: at the heart of the spliceosome
- PMID: 15840809
- PMCID: PMC1370742
- DOI: 10.1261/rna.2220705
Prp8 protein: at the heart of the spliceosome
Abstract
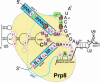
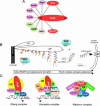
Pre-messenger RNA (pre-mRNA) splicing is a central step in gene expression. Lying between transcription and protein synthesis, pre-mRNA splicing removes sequences (introns) that would otherwise disrupt the coding potential of intron-containing transcripts. This process takes place in the nucleus, catalyzed by a large RNA-protein complex called the spliceosome. Prp8p, one of the largest and most highly conserved of nuclear proteins, occupies a central position in the catalytic core of the spliceosome, and has been implicated in several crucial molecular rearrangements that occur there. Recently, Prp8p has also come under the spotlight for its role in the inherited human disease, Retinitis Pigmentosa.Prp8 is unique, having no obvious homology to other proteins; however, using bioinformatical analysis we reveal the presence of a conserved RNA recognition motif (RRM), an MPN/JAB domain and a putative nuclear localization signal (NLS). Here, we review biochemical and genetical data, mostly related to the human and yeast proteins, that describe Prp8's central role within the spliceosome and its molecular interactions during spliceosome formation, as splicing proceeds, and in post-splicing complexes.
Figures





References
-
- Abovich, N. and Rosbash, M. 1997. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89: 403–412. - PubMed
-
- Achsel, T., Ahrens, K., Brahms, H., Teigelkamp, S., and Luhrmann, R. 1998. The human U5–220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homolog of ribosomal elongation factor EF-2, and a novel WD-40 protein. Mol. Cell. Biol. 18: 6756–6766. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
