Seawi--a sea urchin piwi/argonaute family member is a component of MT-RNP complexes
- PMID: 15840816
- PMCID: PMC1370751
- DOI: 10.1261/rna.7198205
Seawi--a sea urchin piwi/argonaute family member is a component of MT-RNP complexes
Abstract
The piwi/argonaute family of proteins is involved in key developmental processes such as stem cell maintenance and axis specification through molecular mechanisms that may involve RNA silencing. Here we report on the cloning and characterization of the sea urchin piwi/argonaute family member seawi. Seawi is a major component of microtubule-ribonucleoprotein (MT-RNP) complexes isolated from two different species of sea urchin, Strongylocentrotus purpuratus and Paracentrotus lividus. Seawi co-isolates with purified ribosomes, cosediments with 80S ribosomes in sucrose density gradients, and binds microtubules. Seawi possesses the RNA binding motif common to piwi family members and binds P. lividus bep4 mRNA, a transcript that co-isolates with MT-RNP complexes and whose translation product has been shown to play a role in patterning the animal-vegetal axis. Indirect immunofluorescence studies localized seawi to the cortex of unfertilized eggs within granule-like particles, the mitotic spindle during cell division, and the small micromeres where its levels were enriched during the early cleavage stage. Lastly, we discuss how seawi, as a piwi/argonaute family member, may play a fundamentally important role in sea urchin animal-vegetal axis formation and stem cell maintenance.
Figures



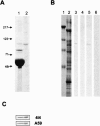



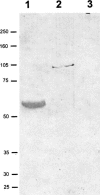


Similar articles
-
Piwi regulates Vasa accumulation during embryogenesis in the sea urchin.Dev Dyn. 2014 Mar;243(3):451-8. doi: 10.1002/dvdy.24096. Dev Dyn. 2014. PMID: 24218044 Free PMC article.
-
Cloning and characterization of HLC-32, a 32-kDa protein component of the sea urchin extraembryonic matrix, the hyaline layer.Dev Biol. 1994 Oct;165(2):556-65. doi: 10.1006/dbio.1994.1275. Dev Biol. 1994. PMID: 7958421
-
Cloning, expression, and localization of a new member of a Paracentrotus lividus cell surface multigene family.Mol Reprod Dev. 1996 May;44(1):36-43. doi: 10.1002/(SICI)1098-2795(199605)44:1<36::AID-MRD4>3.0.CO;2-U. Mol Reprod Dev. 1996. PMID: 8722690
-
Microtubule motors in the early sea urchin embryo.Curr Top Dev Biol. 1992;26:71-91. doi: 10.1016/s0070-2153(08)60441-x. Curr Top Dev Biol. 1992. PMID: 1532933 Review.
-
EML proteins in microtubule regulation and human disease.Biochem Soc Trans. 2016 Oct 15;44(5):1281-1288. doi: 10.1042/BST20160125. Biochem Soc Trans. 2016. PMID: 27911710 Review.
Cited by
-
Senescence and Longevity of Sea Urchins.Genes (Basel). 2020 May 20;11(5):573. doi: 10.3390/genes11050573. Genes (Basel). 2020. PMID: 32443861 Free PMC article.
-
Small RNAs in the animal gonad: guarding genomes and guiding development.Int J Biochem Cell Biol. 2010 Aug;42(8):1334-47. doi: 10.1016/j.biocel.2010.03.005. Epub 2010 Mar 19. Int J Biochem Cell Biol. 2010. PMID: 20227517 Free PMC article. Review.
-
Untangling the web: the diverse functions of the PIWI/piRNA pathway.Mol Reprod Dev. 2013 Aug;80(8):632-64. doi: 10.1002/mrd.22195. Epub 2013 Jun 27. Mol Reprod Dev. 2013. PMID: 23712694 Free PMC article. Review.
-
The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response.Genes Dev. 2007 Dec 15;21(24):3381-94. doi: 10.1101/gad.461107. Genes Dev. 2007. PMID: 18079183 Free PMC article.
-
Maelstrom coordinates microtubule organization during Drosophila oogenesis through interaction with components of the MTOC.Genes Dev. 2011 Nov 15;25(22):2361-73. doi: 10.1101/gad.174110.111. Genes Dev. 2011. PMID: 22085963 Free PMC article.
References
-
- Angerer, L.M. and Angerer, R.C. 2000. Animal–vegetal axis patterning mechanisms in the early sea urchin embryo. Dev. Biol. 218: 1–12. - PubMed
-
- Barbarese, E., Koppel, D.E., Deutscher, M.P., Smith, C.L., Ainger, K., Morgan, F., and Carson, J.H. 1995. Protein translation components are colocalized in granules in oligodendrocytes. J. Cell Sci. 108: 2781–2790. - PubMed
-
- Bonder, E.M., Fishkind, D.J., Cotran, N.M., and Begg, D.A. 1989. The cortical actin-membrane cytoskeleton of unfertilized sea urchin eggs: Analysis of the spatial organization and relationship of filamentous actin, nonfilamentous actin, and egg spectrin. Dev. Biol. 134: 327–341. - PubMed
-
- Brandhorst, B.P. and Klein, W.H. 2002. Molecular patterning along the sea urchin animal–vegetal axis. Int. Rev. Cytol. 213: 183–232. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
