A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies
- PMID: 15840819
- PMCID: PMC1370757
- DOI: 10.1261/rna.2340405
A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies
Abstract
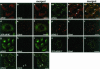
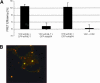
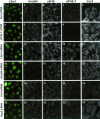
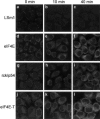
mRNP remodeling events required for the transition of an mRNA from active translation to degradation are currently poorly understood. We identified protein factors potentially involved in this transition, which are present in mammalian P bodies, cytoplasmic foci enriched in 5' --> 3' mRNA degrading enzymes. We demonstrate that human P bodies contain the cap-binding protein eIF4E and the related factor eIF4E-transporter (eIF4E-T), suggesting novel roles for these proteins in targeting mRNAs for 5' --> 3' degradation. Furthermore, fluorescence resonance energy transfer (FRET) studies indicate that eIF4E interacts with eIF4E-T and the putative DEAD box helicase rck/p54 in the P bodies in vivo. RNAi-mediated knockdowns revealed that a subset of P body factors, including eIF4E-T, LSm1, rck/p54, and Ccr4 are required for the accumulation of each other and eIF4E in P bodies. In addition, treatment of HeLa cells with cycloheximide, which inhibits translation, revealed that mRNA is also required for accumulation of mRNA degradation factors in P bodies. In contrast, knockdown of the decapping enzyme Dcp2, which initiates the actual 5' --> 3' mRNA degradation did not abolish P body formation, indicating it first functions after mRNPs have been targeted to these cytoplasmic foci. These data support a model in which mRNPs undergo several successive steps of remodeling and/or 3' trimming until their composition or structural organization promotes their accumulation in P bodies.
Figures


References
-
- Bastiaens, P.I. and Jovin, T.M. 1998. Fluorescence energy transfer microscopy (FRET). In Cell biology: A laboratory handbook (ed. J.E. Celis), Vol. III, pp. 136–146. Academic Press, New York.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
