Structural consequences of the familial amyotrophic lateral sclerosis SOD1 mutant His46Arg
- PMID: 15840828
- PMCID: PMC2253262
- DOI: 10.1110/ps.041256705
Structural consequences of the familial amyotrophic lateral sclerosis SOD1 mutant His46Arg
Abstract
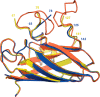
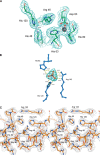
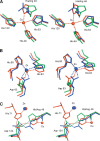
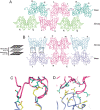
The His46Arg (H46R) mutant of human copper-zinc superoxide dismutase (SOD1) is associated with an unusual, slowly progressing form of familial amyotrophic lateral sclerosis (FALS). Here we describe in detail the crystal structures of pathogenic H46R SOD1 in the Zn-loaded (Zn-H46R) and metal-free (apo-H46R) forms. The Zn-H46R structure demonstrates a novel zinc coordination that involves only three of the usual four liganding residues, His 63, His 80, and Asp 83 together with a water molecule. In addition, the Asp 124 "secondary bridge" between the copper- and zinc-binding sites is disrupted, and the "electrostatic loop" and "zinc loop" elements are largely disordered. The apo-H46R structure exhibits partial disorder in the electrostatic and zinc loop elements in three of the four dimers in the asymmetric unit, while the fourth has ordered loops due to crystal packing interactions. In both structures, nonnative SOD1-SOD1 interactions lead to the formation of higher-order filamentous arrays. The disordered loop elements may increase the likelihood of protein aggregation in vivo, either with other H46R molecules or with other critical cellular components. Importantly, the binding of zinc is not sufficient to prevent the formation of nonnative interactions between pathogenic H46R molecules. The increased tendency to aggregate, even in the presence of Zn, arising from the loss of the secondary bridge is consistent with the observation of an increased abundance of hyaline inclusions in spinal motor neurons and supporting cells in H46R SOD1 transgenic rats.
Figures


References
-
- Aoki, M., Ogasawara, M., Matsubara, Y., Narisawa, K., Nakamura, S., Itoyama, Y., and Abe, K. 1994. Familial amyotrophic-lateral-sclerosis (ALS) in Japan associated with H46R mutation in Cu/Zn superoxide-dismutase gene: A possible new subtype of familial ALS. J. Neurol. Sci. 126 77–83. - PubMed
-
- Arisato, T.O.R., Arata, H., Abe, K., Fukada, K., Sakoda, S., Shimizu, A., Qin, X.H., Izumo, S., Osame, M., and Nakagawa, M. 2003. Clinical and pathological studies of familial amyotrophic lateral sclerosis (FALS) with SOD1 H46R mutation in large Japanese families. Acta Neuropathol. 106 561–568. - PubMed
-
- Banci, L., Felli, I.C., and Kummerle, R. 2002. Direct detection of hydrogen bonds in monomeric superoxide dismutase: Biological implications. Biochemistry 41 2913–2920. - PubMed
-
- Bernstein, F.C., Koetzle, T.F., Williams, G.J.B., Meyer Jr., F.M., Brice, M.D., Rodgers, J.R., Kennard, O., Shimanouchi, T., and Tasumi, M. 1977. The Protein Data Bank: A computer based archival file for macromolecular structures. J. Neurol. Biol. 112 535–542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

