Brain-specific knock-out of hypoxia-inducible factor-1alpha reduces rather than increases hypoxic-ischemic damage
- PMID: 15843612
- PMCID: PMC6724950
- DOI: 10.1523/JNEUROSCI.4555-04.2005
Brain-specific knock-out of hypoxia-inducible factor-1alpha reduces rather than increases hypoxic-ischemic damage
Erratum in
- J Neurosci. 2005 May 11;25(19):1 p following 4888
Abstract
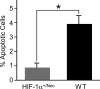
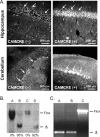
Hypoxia-inducible factor-1alpha (HIF-1alpha) plays an essential role in cellular and systemic O(2) homeostasis by regulating the expression of genes important in glycolysis, erythropoiesis, angiogenesis, and catecholamine metabolism. It is also believed to be a key component of the cellular response to hypoxia and ischemia under pathophysiological conditions, such as stroke. To clarify the function of HIF-1alpha in the brain, we exposed adult mice with late-stage brain deletion of HIF-1alpha to hypoxic injuries. Contrary to expectations, the brains from the HIF-1alpha-deficient mice were protected from hypoxia-induced cell death. These surprising findings suggest that decreasing the level of HIF-1alpha can be neuroprotective. Gene chip expression analysis revealed that, contrary to expectations, the majority of hypoxia-dependent gene-expression changes were unaltered, whereas a specific downregulation of apoptotic genes was observed in the HIF-1alpha-deficient mice. Although the role of HIF-1alpha has been extensively characterized in vitro, in cancer models, and in chronic preconditioning paradigms, this is the first study to evaluate the role of HIF-1alpha in vivo in the brain in response to acute hypoxia/ischemia. Our data suggest, that in acute hypoxia, the neuroprotection found in the HIF-1alpha-deficient mice is mechanistically consistent with a predominant role of HIF-1alpha as proapoptotic and loss of function leads to neuroprotection. Furthermore, our data suggest that functional redundancy develops after excluding HIF-1alpha, leading to the preservation of gene expression regulating the majority of other previously characterized HIF-dependent genes.
Figures


Similar articles
-
Prodeath or prosurvival: two facets of hypoxia inducible factor-1 in perinatal brain injury.Exp Neurol. 2009 Mar;216(1):7-15. doi: 10.1016/j.expneurol.2008.10.016. Epub 2008 Nov 11. Exp Neurol. 2009. PMID: 19041643 Free PMC article. Review.
-
The role and regulation of hypoxia-inducible factor-1alpha expression in brain development and neonatal hypoxic-ischemic brain injury.Brain Res Rev. 2009 Dec 11;62(1):99-108. doi: 10.1016/j.brainresrev.2009.09.006. Epub 2009 Sep 25. Brain Res Rev. 2009. PMID: 19786048 Review.
-
mTOR activates hypoxia-inducible factor-1α and inhibits neuronal apoptosis in the developing rat brain during the early phase after hypoxia-ischemia.Neurosci Lett. 2012 Jan 24;507(2):118-23. doi: 10.1016/j.neulet.2011.11.058. Epub 2011 Dec 8. Neurosci Lett. 2012. PMID: 22178140 Free PMC article.
-
Relationship between HIF-1alpha expression and neuronal apoptosis in neonatal rats with hypoxia-ischemia brain injury.Brain Res. 2007 Nov 14;1180:133-9. doi: 10.1016/j.brainres.2007.08.059. Epub 2007 Sep 5. Brain Res. 2007. PMID: 17920049
-
[Relationship between hypoxia inducible factor 1alpha: expression and neuron apoptosis during hypoxia ischemia brain damage in neonatal rats].Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2007 Dec;21(12):1326-9. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2007. PMID: 18277677 Chinese.
Cited by
-
Group II Metabotropic Glutamate Receptors Reduce Apoptosis and Regulate BDNF and GDNF Levels in Hypoxic-Ischemic Injury in Neonatal Rats.Int J Mol Sci. 2022 Jun 23;23(13):7000. doi: 10.3390/ijms23137000. Int J Mol Sci. 2022. PMID: 35806000 Free PMC article.
-
Ras family small GTPase-mediated neuroprotective signaling in stroke.Cent Nerv Syst Agents Med Chem. 2011 Jun 1;11(2):114-37. doi: 10.2174/187152411796011349. Cent Nerv Syst Agents Med Chem. 2011. PMID: 21521171 Free PMC article. Review.
-
Deferoxamine Preconditioning of Neural-Like Cells Derived from Human Wharton's Jelly Mesenchymal Stem Cells as a Strategy to Promote Their Tolerance and Therapeutic Potential: An In Vitro Study.Cell Mol Neurobiol. 2016 Jul;36(5):689-700. doi: 10.1007/s10571-015-0249-8. Epub 2015 Aug 5. Cell Mol Neurobiol. 2016. PMID: 26242172 Free PMC article.
-
HIF-1 and ventilatory acclimatization to chronic hypoxia.Respir Physiol Neurobiol. 2008 Dec 10;164(1-2):282-7. doi: 10.1016/j.resp.2008.07.017. Respir Physiol Neurobiol. 2008. PMID: 18708172 Free PMC article. Review.
-
Prodeath or prosurvival: two facets of hypoxia inducible factor-1 in perinatal brain injury.Exp Neurol. 2009 Mar;216(1):7-15. doi: 10.1016/j.expneurol.2008.10.016. Epub 2008 Nov 11. Exp Neurol. 2009. PMID: 19041643 Free PMC article. Review.
References
-
- Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley JN, Ried T, Tagle D, Wynshaw-Boris A (1996) Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell 86: 159-171. - PubMed
-
- Berg JT, Breen EC, Fu Z, Mathieu-Costello O, West JB (1998) Alveolar hypoxia increases gene expression of extracellular matrix proteins and platelet-derived growth factor-B in lung parenchyma. Am J Respir Crit Care Med 158: 1920-1928. - PubMed
-
- Bergeron M, Gidday JM, Yu AY, Semenza GL, Ferriero DM, Sharp FR (2000) Role of hypoxia-inducible factor-1 in hypoxia-induced ischemic tolerance in neonatal rat brain. Ann Neurol 48: 285-296. - PubMed
-
- Brusselmans K, Compernolle V, Tjwa M, Wiesener MS, Maxwell PH, Collen D, Carmeliet P (2003) Heterozygous deficiency of hypoxia-inducible factor-2alpha protects mice against pulmonary hypertension and right ventricular dysfunction during prolonged hypoxia. J Clin Invest 111: 1519-1527. - PMC - PubMed
-
- Bunn HF, Poyton RO (1996) Oxygen sensing and molecular adaptation to hypoxia. Physiol Rev 76: 839-885. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
