Relapsing fever spirochetes contain chromosomal genes with unique direct tandemly repeated sequences
- PMID: 15845510
- PMCID: PMC1087331
- DOI: 10.1128/IAI.73.5.3025-3037.2005
Relapsing fever spirochetes contain chromosomal genes with unique direct tandemly repeated sequences
Abstract
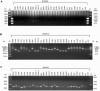
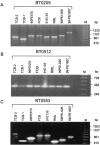
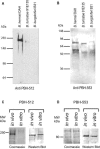
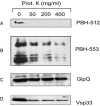
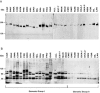
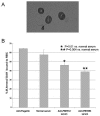
Genome sequencing of the relapsing fever spirochetes Borrelia hermsii and Borrelia turicatae identified three open reading frames (ORFs) on the chromosomes that contained internal, tandemly repeated amino acid sequences that were absent in the Lyme disease spirochete Borrelia burgdorferi. The predicted amino acid sequences of these genes (BH0209, BH0512, and BH0553) have hydrophobic N termini, indicating that these proteins may be secreted. B. hermsii transcribed the three ORFs in vitro, and the BH0512- and BH0553-encoded proteins (PBH-512 and PBH-553) were produced in vitro and in experimentally infected mice. PBH-512 and PBH-553 were on the spirochete's outer surface, and antiserum to these proteins reduced the adherence of B. hermsii to red blood cells. PCR analyses of 28 isolates of B. hermsii and 8 isolates of B. turicatae demonstrated polymorphism in each gene correlated with the number of repeats. Serum samples from relapsing fever patients reacted with recombinant PBH-512 and PBH-553, suggesting that these proteins are produced during human infection. These polymorphic proteins may be involved in the pathogenicity of these relapsing fever spirochetes and provide a mechanism for antigenic heterogeneity within their populations.
Figures


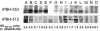
References
-
- Andrade, M. A., C. Perez-Iratxeta, and C. P. Ponting. 2001. Protein repeats: structures, functions, and evolution. J. Struct. Biol. 134:117-131. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

