The Bartonella vinsonii subsp. arupensis immunodominant surface antigen BrpA gene, encoding a 382-kilodalton protein composed of repetitive sequences, is a member of a multigene family conserved among bartonella species
- PMID: 15845521
- PMCID: PMC1087387
- DOI: 10.1128/IAI.73.5.3128-3136.2005
The Bartonella vinsonii subsp. arupensis immunodominant surface antigen BrpA gene, encoding a 382-kilodalton protein composed of repetitive sequences, is a member of a multigene family conserved among bartonella species
Abstract
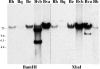
Bartonella proteins that elicit an antibody response during an infection are poorly defined; therefore, to characterize antigens recognized by the host, a Bartonella genomic expression library was screened with serum from an infected mouse. This process led to the discovery of a Bartonella vinsonii subsp. arupensis gene encoding a 382-kDa protein, part of a gene family encoding large proteins, each containing multiple regions of repetitive segments. The genes were termed brpA to -C (bartonella repeat protein) and bore significant similarity to genes encoding the BadA adhesin protein and members of the variably expressed outer membrane protein family of proteins from Bartonella henselae and Bartonella quintana, respectively.
Figures
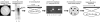



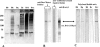

Similar articles
-
The 75-kilodalton antigen of Bartonella bacilliformis is a structural homolog of the cell division protein FtsZ.J Bacteriol. 1997 Jul;179(14):4545-52. doi: 10.1128/jb.179.14.4545-4552.1997. J Bacteriol. 1997. PMID: 9226264 Free PMC article.
-
Production of recombinant Bartonella henselae 17-kDa protein for antibody-capture enzyme-linked immunosorbent assay.Diagn Microbiol Infect Dis. 2006 May;55(1):1-7. doi: 10.1016/j.diagmicrobio.2005.10.020. Epub 2006 Feb 20. Diagn Microbiol Infect Dis. 2006. PMID: 16490335
-
Identification and characterization of an immunodominant 28-kilodalton Coxiella burnetii outer membrane protein specific to isolates associated with acute disease.Infect Immun. 2005 Mar;73(3):1561-7. doi: 10.1128/IAI.73.3.1561-1567.2005. Infect Immun. 2005. PMID: 15731054 Free PMC article.
-
Culture-negative infectious endocarditis caused by Bartonella spp.: 2 case reports and a review of the literature.Diagn Microbiol Infect Dis. 2008 Aug;61(4):476-83. doi: 10.1016/j.diagmicrobio.2008.03.008. Epub 2008 May 1. Diagn Microbiol Infect Dis. 2008. PMID: 18455348 Review.
-
[Bacteria of Bartonella genus: characteristics, pathogenicity factors and methods of molecular-genetic investigation].Mol Gen Mikrobiol Virusol. 2004;(2):3-10. Mol Gen Mikrobiol Virusol. 2004. PMID: 15164714 Review. Russian.
Cited by
-
Analysis of Bartonella adhesin A expression reveals differences between various B. henselae strains.Infect Immun. 2007 Jan;75(1):35-43. doi: 10.1128/IAI.00963-06. Epub 2006 Oct 23. Infect Immun. 2007. PMID: 17060468 Free PMC article.
-
Bartonella quintana variably expressed outer membrane proteins mediate vascular endothelial growth factor secretion but not host cell adherence.Infect Immun. 2006 Sep;74(9):5003-13. doi: 10.1128/IAI.00663-06. Infect Immun. 2006. PMID: 16926391 Free PMC article.
-
Interaction with the host: the role of fibronectin and extracellular matrix proteins in the adhesion of Gram-negative bacteria.Med Microbiol Immunol. 2020 Jun;209(3):277-299. doi: 10.1007/s00430-019-00644-3. Epub 2019 Nov 29. Med Microbiol Immunol. 2020. PMID: 31784893 Free PMC article. Review.
-
Bartonella Species, an Emerging Cause of Blood-Culture-Negative Endocarditis.Clin Microbiol Rev. 2017 Jul;30(3):709-746. doi: 10.1128/CMR.00013-17. Clin Microbiol Rev. 2017. PMID: 28490579 Free PMC article. Review.
-
Characterization of an immunogenic outer membrane autotransporter protein, Arp, of Bartonella henselae.Infect Immun. 2007 Nov;75(11):5255-63. doi: 10.1128/IAI.00533-07. Epub 2007 Sep 4. Infect Immun. 2007. PMID: 17785470 Free PMC article.
References
-
- Alsmark, C. M., A. C. Frank, E. O. Karlberg, B. A. Legault, D. H. Ardell, B. Canback, A. S. Eriksson, A. K. Naslund, S. A. Handley, M. Huvet, B. La Scola, M. Holmberg, and S. G. Andersson. 2004. The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae. Proc. Natl. Acad. Sci. USA 101:9716-9721. - PMC - PubMed
-
- Bai, Y., M. Y. Kosoy, G. O. Maupin, K. R. Tsuchiya, and K. L. Gage. 2002. Genetic and ecologic characteristics of Bartonella communities in rodents in southern China. Am. J. Trop. Med. Hyg. 66:622-627. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

