Virus-inducible reporter genes as a tool for detecting and quantifying influenza A virus replication
- PMID: 15847914
- PMCID: PMC1698269
- DOI: 10.1016/j.jviromet.2005.01.016
Virus-inducible reporter genes as a tool for detecting and quantifying influenza A virus replication
Abstract
The use of influenza A virus-inducible reporter gene segments in detecting influenza A virus replication was investigated. The RNA polymerase I promoter/terminator cassette was used to express RNA transcripts encoding green fluorescence protein or firefly luciferase flanked by the untranslated regions of the influenza A/WSN/33 nucleoprotein (NP) segment. Reporter gene activity was detected after reconstitution of the influenza A virus polymerase complex from cDNA or after virus infection, and was influenza A virus-specific. Reporter gene activity could be detected as early as 6 h post-infection and was virus dose-dependent. Inhibitory effects of antibodies or amantadine could be detected and quantified rapidly, providing a means of not only identifying influenza A virus-specific replication, but also of determining the antigenic subtype as well as antiviral drug susceptibility. Induction of virus-specific reporter genes provides a rapid, sensitive method for detecting virus replication, quantifying virus titers and assessing antiviral sensitivity as well as antigenic subtype.
Figures
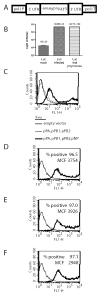
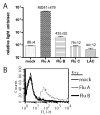
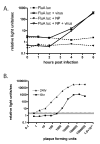
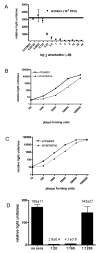
References
-
- Azzeh M, Flick R, Hobom G. Functional analysis of the influenza A virus cRNA promoter and construction of an ambisense transcription system. Virology. 2001;289(2):400–10. - PubMed
-
- Catchpole AP, Mingay LJ, Fodor E, Brownlee GG. Alternative base pairs attenuate influenza A virus when introduced into the duplex region of the conserved viral RNA promoter of either the NS or the PA gene. J Gen Virol. 2003;84(3):507–515. - PubMed
-
- Crescenzo-Chaigne B, Naffakh N, van der Werf S. Comparative Analysis of the Ability of the Polymerase Complexes of Influenza Viruses Type A, B and C to Assemble into Functional RNPs that Allow Expression and Replication of Heterotypic Model RNA Templates In Vivo. Virology. 1999;265(2):342–353. - PubMed
-
- Ellis JS, Zambon MC. Molecular diagnosis of influenza. Rev Med Virol. 2002;12(6):375–89. - PubMed
-
- Enami M, Sharma G, Benham C, Palese P. An influenza virus containing nine different RNA segments. Virology. 1991;185(1):291–8. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

