Recombinant severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein forms a dimer through its C-terminal domain
- PMID: 15849181
- PMCID: PMC8008353
- DOI: 10.1074/jbc.M501015200
Recombinant severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein forms a dimer through its C-terminal domain
Abstract
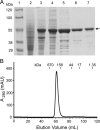
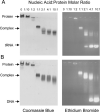
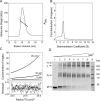
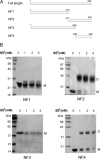
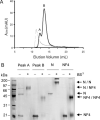
The causative agent of severe acute respiratory syndrome (SARS) is the SARS-associated coronavirus, SARS-CoV. The viral nucleocapsid (N) protein plays an essential role in viral RNA packaging. In this study, recombinant SARS-CoV N protein was shown to be dimeric by analytical ultracentrifugation, size exclusion chromatography coupled with light scattering, and chemical cross-linking. Dimeric N proteins self-associate into tetramers and higher molecular weight oligomers at high concentrations. The dimerization domain of N was mapped through studies of the oligomeric states of several truncated mutants. Although mutants consisting of residues 1-210 and 1-284 fold as monomers, constructs consisting of residues 211-422 and 285-422 efficiently form dimers. When in excess, the truncated construct 285-422 inhibits the homodimerization of full-length N protein by forming a heterodimer with the full-length N protein. These results suggest that the N protein oligomerization involves the C-terminal residues 285-422, and this region is a good target for mutagenic studies to disrupt N protein self-association and virion assembly.
Figures
Similar articles
-
Carboxyl terminus of severe acute respiratory syndrome coronavirus nucleocapsid protein: self-association analysis and nucleic acid binding characterization.Biochemistry. 2006 Oct 3;45(39):11827-35. doi: 10.1021/bi0609319. Biochemistry. 2006. PMID: 17002283
-
Biochemical and immunological studies of nucleocapsid proteins of severe acute respiratory syndrome and 229E human coronaviruses.Proteomics. 2005 Mar;5(4):925-37. doi: 10.1002/pmic.200401204. Proteomics. 2005. PMID: 15759315 Free PMC article.
-
Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between corona- and arteriviridae.J Biol Chem. 2006 Jun 23;281(25):17134-17139. doi: 10.1074/jbc.M602107200. Epub 2006 Apr 20. J Biol Chem. 2006. PMID: 16627473 Free PMC article.
-
SR-rich motif plays a pivotal role in recombinant SARS coronavirus nucleocapsid protein multimerization.Biochemistry. 2005 Nov 22;44(46):15351-8. doi: 10.1021/bi051122c. Biochemistry. 2005. PMID: 16285739
-
The SARS coronavirus nucleocapsid protein--forms and functions.Antiviral Res. 2014 Mar;103:39-50. doi: 10.1016/j.antiviral.2013.12.009. Epub 2014 Jan 11. Antiviral Res. 2014. PMID: 24418573 Free PMC article. Review.
Cited by
-
Interaction of coronavirus nucleocapsid protein with the 5'- and 3'-ends of the coronavirus genome is involved in genome circularization and negative-strand RNA synthesis.FEBS J. 2019 Aug;286(16):3222-3239. doi: 10.1111/febs.14863. Epub 2019 May 8. FEBS J. 2019. PMID: 31034708 Free PMC article.
-
mRNA display design of fibronectin-based intrabodies that detect and inhibit severe acute respiratory syndrome coronavirus nucleocapsid protein.J Biol Chem. 2009 Jun 26;284(26):17512-20. doi: 10.1074/jbc.M901547200. Epub 2009 Apr 13. J Biol Chem. 2009. PMID: 19364769 Free PMC article.
-
A conserved oligomerization domain in the disordered linker of coronavirus nucleocapsid proteins.Sci Adv. 2023 Apr 5;9(14):eadg6473. doi: 10.1126/sciadv.adg6473. Epub 2023 Apr 5. Sci Adv. 2023. PMID: 37018390 Free PMC article.
-
The dimer interface of the SARS coronavirus nucleocapsid protein adapts a porcine respiratory and reproductive syndrome virus-like structure.FEBS Lett. 2005 Oct 24;579(25):5663-8. doi: 10.1016/j.febslet.2005.09.038. Epub 2005 Sep 30. FEBS Lett. 2005. PMID: 16214138 Free PMC article.
-
Epitope mapping and cellular localization of swine acute diarrhea syndrome coronavirus nucleocapsid protein using a novel monoclonal antibody.Virus Res. 2019 Nov;273:197752. doi: 10.1016/j.virusres.2019.197752. Epub 2019 Sep 10. Virus Res. 2019. PMID: 31518629 Free PMC article.
References
-
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. Science. 2003;300:1399–1404. - PubMed
-
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Science. 2003;300:1394–1399. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

