High frequency trans-splicing in a cell line producing spliced and polyadenylated RNA polymerase I transcripts from an rDNA-myc chimeric gene
- PMID: 15849319
- PMCID: PMC1084326
- DOI: 10.1093/nar/gki530
High frequency trans-splicing in a cell line producing spliced and polyadenylated RNA polymerase I transcripts from an rDNA-myc chimeric gene
Abstract
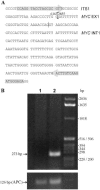
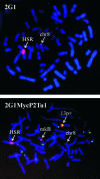
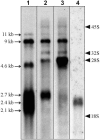
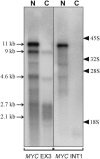
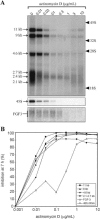
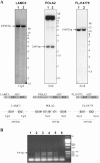
The 2G1MycP2Tu1 cell line was obtained following transfection of human colon carcinoma cells from the SW613-S cell line with a plasmid carrying a genomic copy of the human MYC gene. 2G1MycP2Tu1 cells produce MYC mRNAs and proteins of abnormal size. In order to analyze the structure of these abnormal products, a cDNA library constructed using RNA isolated from these cells was screened with a MYC probe. Fifty clones were studied by DNA sequencing. The results indicated that a truncated copy of the MYC gene had integrated into an rDNA transcription unit in 2G1MycP2Tu1 cells. This was confirmed by northern blot analysis, PCR amplification on genomic DNA and fluorescent in situ hybridization (FISH) experiments on metaphase chromosomes. 2G1MycP2Tu1 cells produce hybrid rRNA-MYC RNA molecules that are polyadenylated and processed by splicing reactions involving natural and cryptic splice sites. These transcripts are synthesized by RNA polymerase I, as confirmed by actinomycin D sensitivity experiments, suggesting that 3' end processing and splicing are uncoupled from transcription in this case. 2G1MycP2Tu1 cells also produce another type of chimeric mRNAs consisting of correctly spliced exons 2 and 3 of the MYC gene fused to one or more extraneous 5' exons by proper splicing to the acceptor sites of MYC exon 2. These foreign exons belong to 33 different genes, which are located on 14 different chromosomes. These observations and the results of FISH and Southern blotting experiments lead us to conclude that trans-splicing events occur at high frequency in 2G1MycP2Tu1 cells.
Figures


Similar articles
-
A candidate chimeric mammalian mRNA transcript is derived from distinct chromosomes and is associated with nonconsensus splice junction motifs.DNA Cell Biol. 2003 May;22(5):303-15. doi: 10.1089/104454903322216653. DNA Cell Biol. 2003. PMID: 12941158
-
Natural trans-spliced mRNAs are generated from the human estrogen receptor-alpha (hER alpha) gene.J Biol Chem. 2002 Jul 19;277(29):26244-51. doi: 10.1074/jbc.M203513200. Epub 2002 May 13. J Biol Chem. 2002. PMID: 12011094
-
Multiple transcription initiation sites, alternative splicing, and differential polyadenylation contribute to the complexity of human neurofibromatosis 2 transcripts.Genomics. 2002 Jan;79(1):63-76. doi: 10.1006/geno.2001.6672. Genomics. 2002. PMID: 11827459
-
mRNA splicing in trypanosomes.Int J Med Microbiol. 2012 Oct;302(4-5):221-4. doi: 10.1016/j.ijmm.2012.07.004. Epub 2012 Sep 7. Int J Med Microbiol. 2012. PMID: 22964417 Review.
-
Intergenic mRNAs. Minor gene products or tools of diversity?Histol Histopathol. 2002 Apr;17(2):677-82. doi: 10.14670/HH-17.677. Histol Histopathol. 2002. PMID: 11962767 Review.
Cited by
-
Novel domain combinations in proteins encoded by chimeric transcripts.Bioinformatics. 2012 Jun 15;28(12):i67-74. doi: 10.1093/bioinformatics/bts216. Bioinformatics. 2012. PMID: 22689780 Free PMC article.
-
Nucleocytoplasmic traffic of CPEB1 and accumulation in Crm1 nucleolar bodies.Mol Biol Cell. 2009 Jan;20(1):176-87. doi: 10.1091/mbc.e08-09-0904. Epub 2008 Oct 15. Mol Biol Cell. 2009. PMID: 18923137 Free PMC article.
-
Loss of functional OPA1 unbalances redox state: implications in dominant optic atrophy pathogenesis.Ann Clin Transl Neurol. 2016 Apr 25;3(6):408-21. doi: 10.1002/acn3.305. eCollection 2016 Jun. Ann Clin Transl Neurol. 2016. PMID: 27547769 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions

