Evidence of selection for low cognate amino acid bias in amino acid biosynthetic enzymes
- PMID: 15853887
- PMCID: PMC1839009
- DOI: 10.1111/j.1365-2958.2005.04566.x
Evidence of selection for low cognate amino acid bias in amino acid biosynthetic enzymes
Abstract
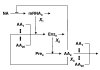
If the enzymes responsible for biosynthesis of a given amino acid are repressed and the cognate amino acid pool suddenly depleted, then derepression of these enzymes and replenishment of the pool would be problematic, if the enzymes were largely composed of the cognate amino acid. In the proverbial "Catch 22", cells would lack the necessary enzymes to make the amino acid, and they would lack the necessary amino acid to make the needed enzymes. Based on this scenario, we hypothesize that evolution would lead to the selection of amino acid biosynthetic enzymes that have a relatively low content of their cognate amino acid. We call this the "cognate bias hypothesis". Here we test several implications of this hypothesis directly using data from the proteome of Escherichia coli. Several lines of evidence show that low cognate bias is evident in 15 of the 20 amino acid biosynthetic pathways. Comparison with closely related Salmonella typhimurium shows similar results. Comparison with more distantly related Bacillus subtilis shows general similarities as well as significant differences in the detailed profiles of cognate bias. Thus, selection for low cognate bias plays a significant role in shaping the amino acid composition for a large class of cellular proteins.
Figures



References
-
- Baudouin-Cornu P, Surdin-Kerjan Y, Marliere P, Thomas D. Molecular evolution of protein atomic composition. Science. 2001;293::297–300. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

