Measuring chromosome dynamics on different time scales using resolvases with varying half-lives
- PMID: 15853889
- PMCID: PMC1373788
- DOI: 10.1111/j.1365-2958.2005.04588.x
Measuring chromosome dynamics on different time scales using resolvases with varying half-lives
Abstract
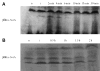
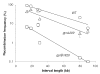
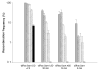
The bacterial chromosome is organized into multiple independent domains, each capable of constraining the plectonemic negative supercoil energy introduced by DNA gyrase. Different experimental approaches have estimated the number of domains to be between 40 and 150. The site-specific resolution systems of closely related transposons Tn3 and gammadelta are valuable tools for measuring supercoil diffusion and analysing bacterial chromosome dynamics in vivo. Once made, the wild-type resolvase persists in cells for time periods greater than the cell doubling time. To examine chromosome dynamics over shorter time frames that are more closely tuned to processes like inducible transcription, we constructed a set of resolvases with cellular half-lives ranging from less than 5 min to 30 min. Analysing chromosomes on different time scales shows domain structure to be dynamic. Rather than the 150 domains detected with the Tn3 resolvase, wild-type cells measured over a 10 min time span have more than 400 domains per genome equivalent, and some gyrase mutants exceed 1000.
Figures


Similar articles
-
Rates of gyrase supercoiling and transcription elongation control supercoil density in a bacterial chromosome.PLoS Genet. 2012;8(8):e1002845. doi: 10.1371/journal.pgen.1002845. Epub 2012 Aug 16. PLoS Genet. 2012. PMID: 22916023 Free PMC article.
-
Transcription-induced barriers to supercoil diffusion in the Salmonella typhimurium chromosome.Proc Natl Acad Sci U S A. 2004 Mar 9;101(10):3398-403. doi: 10.1073/pnas.0307550101. Epub 2004 Mar 1. Proc Natl Acad Sci U S A. 2004. PMID: 14993611 Free PMC article.
-
Measuring In Vivo Supercoil Dynamics and Transcription Elongation Rates in Bacterial Chromosomes.Methods Mol Biol. 2017;1624:17-27. doi: 10.1007/978-1-4939-7098-8_2. Methods Mol Biol. 2017. PMID: 28842872
-
Catalysis of site-specific recombination by Tn3 resolvase.Biochem Soc Trans. 2010 Apr;38(2):417-21. doi: 10.1042/BST0380417. Biochem Soc Trans. 2010. PMID: 20298194 Review.
-
RNA polymerase: chromosome domain boundary maker and regulator of supercoil density.Curr Opin Microbiol. 2014 Dec;22:138-43. doi: 10.1016/j.mib.2014.10.002. Curr Opin Microbiol. 2014. PMID: 25460807 Free PMC article. Review.
Cited by
-
Immunity of replicating Mu to self-integration: a novel mechanism employing MuB protein.Mob DNA. 2010 Feb 1;1(1):8. doi: 10.1186/1759-8753-1-8. Mob DNA. 2010. PMID: 20226074 Free PMC article.
-
The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation.Mol Cell. 2011 Oct 21;44(2):252-64. doi: 10.1016/j.molcel.2011.09.010. Mol Cell. 2011. PMID: 22017872 Free PMC article.
-
Bacterial chromosome organization and segregation.Annu Rev Cell Dev Biol. 2015;31:171-99. doi: 10.1146/annurev-cellbio-100814-125211. Annu Rev Cell Dev Biol. 2015. PMID: 26566111 Free PMC article. Review.
-
New approaches to understanding the spatial organization of bacterial genomes.Curr Opin Microbiol. 2014 Dec;22:15-21. doi: 10.1016/j.mib.2014.09.014. Epub 2014 Oct 7. Curr Opin Microbiol. 2014. PMID: 25305533 Free PMC article. Review.
-
Species-specific supercoil dynamics of the bacterial nucleoid.Biophys Rev. 2016 Nov;8(Suppl 1):113-121. doi: 10.1007/s12551-016-0207-9. Epub 2016 Jul 20. Biophys Rev. 2016. PMID: 28510215 Free PMC article.
References
-
- Benjamin KR, Abola AP, Kanaar R, Cozzarelli NR. Contributions of supercoiling to Tn3 resolvase and phage Mu Gin site-specific recombination. J Mol Biol. 1996;256:50–65. - PubMed
-
- den Blaauwen T, Lindqvist A, Lowe J, Nanninga N. Distribution of the Escherichia coli structural maintenance of chromosomes (SMC)-like protein MukB in the cell. Mol Microbiol. 2001;42:1179–1188. - PubMed
-
- Bliska JB, Cozzarelli NR. Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J Mol Biol. 1987;194:205–218. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

