Some statistical properties of regulatory DNA sequences, and their use in predicting regulatory regions in the Drosophila genome: the fluffy-tail test
- PMID: 15857505
- PMCID: PMC1127108
- DOI: 10.1186/1471-2105-6-109
Some statistical properties of regulatory DNA sequences, and their use in predicting regulatory regions in the Drosophila genome: the fluffy-tail test
Abstract
Background: This paper addresses the problem of recognising DNA cis-regulatory modules which are located far from genes. Experimental procedures for this are slow and costly, and computational methods are hard, because they lack positional information.
Results: We present a novel statistical method, the "fluffy-tail test", to recognise regulatory DNA. We exploit one of the basic informational properties of regulatory DNA: abundance of over-represented transcription factor binding site (TFBS) motifs, although we do not look for specific TFBS motifs, per se . Though overrepresentation of TFBS motifs in regulatory DNA has been intensively exploited by many algorithms, it is still a difficult problem to distinguish regulatory from other genomic DNA.
Conclusion: We show that, in the data used, our method is able to distinguish cis-regulatory modules by exploiting statistical differences between the probability distributions of similar words in regulatory and other DNA. The potential application of our method includes annotation of new genomic sequences and motif discovery.
Figures
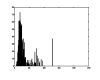
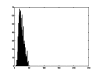
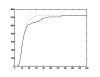

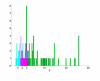
References
-
- Yuh C, Bolouri H, Davidson EH. Cis-regulatory logic in the endo 16 gene: switching from a specification to a differentiation mode of control. Development. 2001;128:617–29. - PubMed
-
- Davidson EH. Genomic Regulatory Systems. Academic Press; 2001.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

