Amino acid 36 in the human immunodeficiency virus type 1 gp41 ectodomain controls fusogenic activity: implications for the molecular mechanism of viral escape from a fusion inhibitor
- PMID: 15857986
- PMCID: PMC1091722
- DOI: 10.1128/JVI.79.10.5996-6004.2005
Amino acid 36 in the human immunodeficiency virus type 1 gp41 ectodomain controls fusogenic activity: implications for the molecular mechanism of viral escape from a fusion inhibitor
Abstract
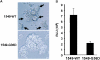
We have previously described a human immunodeficiency virus type 1 (HIV-1) proviral clone, pL2, derived from defective viral particles with higher fusogenicity than the prototypic NL4-3 virus. In this study, we attempted to determine the region that confers the enhanced fusion activity by creating envelope recombinants between pL2 and pNL4-3, as well as point mutants based on pNL4-3. The results indicate that amino acid 36 of gp41 is key for the fusogenic activity and infectivity enhancement and that glycine 36 (36G) of gp41 in pL2 is conserved in nearly all HIV-1 isolates except for pNL4-3. The mutation 36G-->D in a primary-isolate-derived Env decreased syncytium-forming activity and infectivity. The assays for cell-cell fusion and viral binding suggested that the enhanced fusion mediated by the 36D-->G mutation is not due to increased binding efficiency but is directly due to actual enhancement of viral fusion activity. Interestingly, this amino acid position is exactly equivalent to that at which the mutation of HIV-1 isolates that have escaped from a fusion inhibitor, enfuvirtide (T-20), has been frequently observed. The correlation between these previous findings and our findings was suggested by structural analysis. Our finding, therefore, has implications for a molecular basis of the viral escape from this drug.
Figures



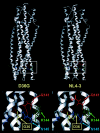

Similar articles
-
Mutations of conserved glycine residues within the membrane-spanning domain of human immunodeficiency virus type 1 gp41 can inhibit membrane fusion and incorporation of Env onto virions.Jpn J Infect Dis. 2006 Apr;59(2):77-84. Jpn J Infect Dis. 2006. PMID: 16632906
-
Role of envelope processing and gp41 membrane spanning domain in the formation of human immunodeficiency virus type 1 (HIV-1) fusion-competent envelope glycoprotein complex.Virus Res. 2007 Mar;124(1-2):103-12. doi: 10.1016/j.virusres.2006.10.009. Epub 2006 Nov 28. Virus Res. 2007. PMID: 17129629
-
The effect of the membrane-proximal tyrosine-based sorting signal of HIV-1 gp41 on viral infectivity depends on sequences within gp120.Virology. 2006 Oct 25;354(2):316-27. doi: 10.1016/j.virol.2006.06.023. Epub 2006 Aug 14. Virology. 2006. PMID: 16905171
-
Conserved residues in the coiled-coil pocket of human immunodeficiency virus type 1 gp41 are essential for viral replication and interhelical interaction.Virology. 2004 Nov 24;329(2):319-27. doi: 10.1016/j.virol.2004.08.025. Virology. 2004. PMID: 15518811
-
Protein design of a bacterially expressed HIV-1 gp41 fusion inhibitor.Biochemistry. 2007 Apr 10;46(14):4360-9. doi: 10.1021/bi7001289. Epub 2007 Mar 20. Biochemistry. 2007. PMID: 17371053
Cited by
-
Highly conserved configuration of catalytic amino acid residues among calicivirus-encoded proteases.J Virol. 2007 Jul;81(13):6798-806. doi: 10.1128/JVI.02840-06. Epub 2007 Apr 25. J Virol. 2007. PMID: 17459935 Free PMC article.
-
Impact of amino acid substitutions in the V2 and C2 regions of human immunodeficiency virus type 1 CRF01_AE envelope glycoprotein gp120 on viral neutralization susceptibility to broadly neutralizing antibodies specific for the CD4 binding site.Retrovirology. 2014 Apr 23;11:32. doi: 10.1186/1742-4690-11-32. Retrovirology. 2014. PMID: 24758333 Free PMC article.
-
Identification and characterization of potent small molecule inhibitor of hemorrhagic fever New World arenaviruses.Antiviral Res. 2006 Feb;69(2):86-97. doi: 10.1016/j.antiviral.2005.10.008. Epub 2005 Nov 28. Antiviral Res. 2006. PMID: 16343651 Free PMC article.
-
SC29EK, a peptide fusion inhibitor with enhanced alpha-helicity, inhibits replication of human immunodeficiency virus type 1 mutants resistant to enfuvirtide.Antimicrob Agents Chemother. 2009 Mar;53(3):1013-8. doi: 10.1128/AAC.01211-08. Epub 2008 Dec 29. Antimicrob Agents Chemother. 2009. PMID: 19114674 Free PMC article.
-
Identification of an Antiretroviral Small Molecule That Appears To Be a Host-Targeting Inhibitor of HIV-1 Assembly.J Virol. 2021 Jan 13;95(3):e00883-20. doi: 10.1128/JVI.00883-20. Print 2021 Jan 13. J Virol. 2021. PMID: 33148797 Free PMC article.
References
-
- Baker, D., and A. Sali. 2001. Protein structure prediction and structural genomics. Science 294:93-96. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

