Carboxyl-proximal regions of reovirus nonstructural protein muNS necessary and sufficient for forming factory-like inclusions
- PMID: 15858004
- PMCID: PMC1091696
- DOI: 10.1128/JVI.79.10.6194-6206.2005
Carboxyl-proximal regions of reovirus nonstructural protein muNS necessary and sufficient for forming factory-like inclusions
Abstract
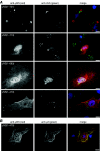
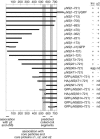
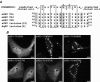
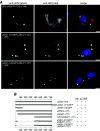
Mammalian orthoreoviruses are believed to replicate in distinctive, cytoplasmic inclusion bodies, commonly called viral factories or viroplasms. The viral nonstructural protein muNS has been implicated in forming the matrix of these structures, as well as in recruiting other components to them for putative roles in genome replication and particle assembly. In this study, we sought to identify the regions of muNS that are involved in forming factory-like inclusions in transfected cells in the absence of infection or other viral proteins. Sequences in the carboxyl-terminal one-third of the 721-residue muNS protein were linked to this activity. Deletion of as few as eight residues from the carboxyl terminus of muNS resulted in loss of inclusion formation, suggesting that some portion of these residues is required for the phenotype. A region spanning residues 471 to 721 of muNS was the smallest one shown to be sufficient for forming factory-like inclusions. The region from positions 471 to 721 (471-721 region) includes both of two previously predicted coiled-coil segments in muNS, suggesting that one or both of these segments may also be required for inclusion formation. Deletion of the more amino-terminal one of the two predicted coiled-coil segments from the 471-721 region resulted in loss of the phenotype, although replacement of this segment with Aequorea victoria green fluorescent protein, which is known to weakly dimerize, largely restored inclusion formation. Sequences between the two predicted coiled-coil segments were also required for forming factory-like inclusions, and mutation of either one His residue (His570) or one Cys residue (Cys572) within these sequences disrupted the phenotype. The His and Cys residues are part of a small consensus motif that is conserved across muNS homologs from avian orthoreoviruses and aquareoviruses, suggesting this motif may have a common function in these related viruses. The inclusion-forming 471-721 region of muNS was shown to provide a useful platform for the presentation of peptides for studies of protein-protein association through colocalization to factory-like inclusions in transfected cells.
Figures



References
-
- Antczak, J. B., and W. K. Joklik. 1992. Reovirus genome segment assortment into progeny genomes studied by the use of monoclonal antibodies directed against reovirus proteins. Virology 187:760-776. - PubMed
-
- Attoui, H., Q. Fang, F. M. Jaafar, J. F. Cantaloube, P. Biagini, P. De Micco, and X. De Lamballerie. 2002. Common evolutionary origin of aquareoviruses and orthoreoviruses revealed by genome characterization of golden shiner reovirus, grass carp reovirus, striped bass reovirus and golden ide reovirus (genus Aquareovirus, family Reoviridae). J. Gen. Virol. 83:1941-1951. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

