Relocalization of the Mre11-Rad50-Nbs1 complex by the adenovirus E4 ORF3 protein is required for viral replication
- PMID: 15858005
- PMCID: PMC1091726
- DOI: 10.1128/JVI.79.10.6207-6215.2005
Relocalization of the Mre11-Rad50-Nbs1 complex by the adenovirus E4 ORF3 protein is required for viral replication
Abstract
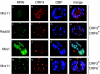
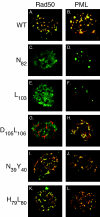
Adenovirus replication is controlled by the relocalization or modification of nuclear protein complexes, including promyelocytic leukemia protein (PML) nuclear domains and the Mre11-Rad50-Nbs1 (MRN) DNA damage machinery. In this study, we demonstrated that the E4 ORF3 protein effects the relocalization of both PML and MRN proteins to similar structures within the nucleus at early times after infection. These proteins colocalize with E4 ORF3. Through the analysis of specific viral mutants, we found a direct correlation between MRN reorganization at early times after infection and the establishment of viral DNA replication domains. Further, the reorganization of MRN components may be uncoupled from the ability of E4 ORF3 to rearrange PML. At later stages of infection, components of the MRN complex disperse within the nucleus, Nbs1 is found within viral replication centers, Rad50 remains localized with E4 ORF3, and Mre11 is degraded. The importance of viral regulation of the MRN complex is underscored by the complementation of E4 mutant viruses in cells that lack Mre11 or Nbs1 activity. These results illustrate the importance of nuclear organization in virus growth and suggest that E4 ORF3 regulates activities in both PML nuclear bodies and the MRN complex to stimulate the viral replication program.
Figures



References
-
- Boyer, J., K. Rohleder, and G. Ketner. 1999. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology 263:307-312. - PubMed
-
- Carbone, R., M. Pearson, S. Minucci, and P. Pelicci. 2002. PML NBs associate with the hMre11 complex and p53 at sites of irradiation induced DNA damage. Oncogene 21:1633-1640. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

