Epitope-mapping studies define two major neutralization sites on the vaccinia virus extracellular enveloped virus glycoprotein B5R
- PMID: 15858010
- PMCID: PMC1091701
- DOI: 10.1128/JVI.79.10.6260-6271.2005
Epitope-mapping studies define two major neutralization sites on the vaccinia virus extracellular enveloped virus glycoprotein B5R
Abstract
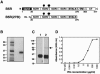
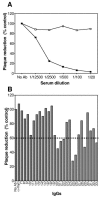
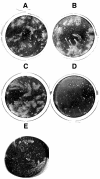
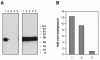
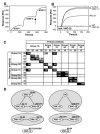
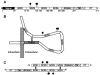
Vaccinia extracellular enveloped virus (EEV) is critical for cell-to-cell and long-range virus spread both in vitro and in vivo. The B5R gene encodes an EEV-specific type I membrane protein that is essential for efficient EEV formation. The majority of the B5R ectodomain consists of four domains with homology to short consensus repeat domains followed by a stalk. Previous studies have shown that polyclonal antibodies raised against the B5R ectodomain inhibit EEV infection. In this study, our goal was to elucidate the antigenic structure of B5R and relate this to its function. To do this, we produced multimilligram quantities of vaccinia virus B5R as a soluble protein [B5R(275t)] using a baculovirus expression system. We then selected and characterized a panel of 26 monoclonal antibodies (MAbs) that recognize B5R(275t). Five of these MAbs neutralized EEV and inhibited comet formation. Two other MAbs were able only to neutralize EEV, while five others were able only to inhibit comet formation. This suggests that the EEV neutralization and comet inhibition assays measure different viral functions and that at least two different antigenic sites on B5R are important for these activities. We further characterized the MAbs and the antigenic structure of B5R(275t) by peptide mapping and by reciprocal MAb blocking studies using biosensor analysis. The epitopes recognized by neutralizing MAbs were localized to SCR1-SCR2 and/or the stalk of B5R(275t). Furthermore, the peptide and blocking data support the concept that SCR1 and the stalk may be in juxtaposition and may be part of the same functional domain.
Figures




References
-
- Appleyard, G., A. J. Hapel, and E. A. Boulter. 1971. An antigenic difference between intracellular and extracellular rabbitpox virus. J. Gen. Virol. 13:9-17. - PubMed
-
- Aslam, M., and S. J. Perkins. 2001. Folded-back solution structure of monomeric factor H of human complement by synchrotron X-ray and neutron scattering, analytical ultracentrifugation and constrained molecular modelling. J. Mol. Biol. 309:1117-1138. - PubMed
-
- Bell, E., M. Shamim, J. C. Whitbeck, G. Sfyroera, J. D. Lambris, and S. N. Isaacs. 2004. Antibodies against the extracellular enveloped virus B5R protein are mainly responsible for the EEV neutralizing capacity of vaccinia immune globulin. Virology 325:425-431. - PubMed
-
- Boulter, E. A., and G. Appleyard. 1973. Differences between extracellular and intracellular forms of poxvirus and their implications. Prog. Med. Virol. 16:86-108. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

