Compartmentalization of hepatitis C virus genotypes between plasma and peripheral blood mononuclear cells
- PMID: 15858018
- PMCID: PMC1091708
- DOI: 10.1128/JVI.79.10.6349-6357.2005
Compartmentalization of hepatitis C virus genotypes between plasma and peripheral blood mononuclear cells
Abstract
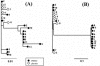
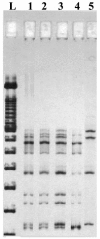
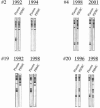
Differences in hepatitis C virus (HCV) variants of the highly conserved 5' untranslated region (UTR) have been observed between plasma and peripheral blood mononuclear cells (PBMC). The prevalence and the mechanisms of this compartmentalization are unknown. Plasma and PBMC HCV variants were compared by single-strand conformation polymorphism (SSCP) and by cloning or by genotyping with a line probe assay (LiPA) in 116 chronically infected patients, including 44 liver transplant recipients. SSCP patterns differed between compartments in 43/109 analyzable patients (39%). Differences were significantly more frequent in patients with transplants (21/38 [55%] versus 22/71 [31%]; P < 0.01) and in those who acquired HCV through multiple transfusions before 1991 (15/20; 75%) or through drug injection (16/31; 52%) than in those infected through an unknown route (7/29; 24%) or through a single transfusion (5/29; 17%; P < 0.001). Cloning of the 5' UTR, LiPA analysis, and nonstructural region 5B sequencing revealed different genotypes in the two compartments from 10 patients (9%). In nine patients, the genotype detected in PBMC was not detected in plasma and was weak or undetectable in the liver in three cases. This genotypic compartmentalization persisted for years in three patients and after liver transplantation in two. The present study shows that a significant proportion of HCV-infected subjects harbor in their PBMC highly divergent variants which were likely acquired through superinfections.
Figures

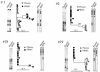
References
-
- Bain, C., A. Fatmi, F. Zoulim, J. P. Zarski, C. Trepo, and G. Inchauspe. 2001. Impaired allostimulatory function of dendritic cells in chronic hepatitis C infection. Gastroenterology 120:512-524. - PubMed
-
- Bedossa, P., T. Poynard, et al. 1996. An algorithm for the grading of activity in chronic hepatitis C. 24:289-293. - PubMed
-
- Boisvert, J., X. S. He, R. Cheung, E. B. Keeffe, T. Wright, and H. B. Greenberg. 2001. Quantitative analysis of hepatitis C virus in peripheral blood and liver: replication detected only in liver. J. Infect. Dis. 184:827-835. - PubMed
-
- Bronowicki, J., M. Loriot, V. Thiers, Y. Grignon, A. Zignego, and C. Brechot. 1999. Hepatitis C virus persistence in human hematopoietic cells injected into SCID mice. Hepatology 28:211-218. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

