Molecular classification of renal tumors by gene expression profiling
- PMID: 15858144
- PMCID: PMC1867531
- DOI: 10.1016/S1525-1578(10)60547-8
Molecular classification of renal tumors by gene expression profiling
Abstract
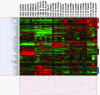
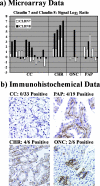
Renal tumor classification is important because histopathological subtypes are associated with distinct clinical behavior. However, diagnosis is difficult because tumor subtypes have overlapping microscopic characteristics. Therefore, ancillary methods are needed to optimize classification. We used oligonucleotide microarrays to analyze 31 adult renal tumors, including clear cell renal cell carcinoma (RCC), papillary RCC, chromophobe RCC, oncocytoma, and angiomyolipoma. Expression profiles correlated with histopathology; unsupervised algorithms clustered 30 of 31 tumors according to appropriate diagnostic subtypes while supervised analyses identified significant, subtype-specific expression markers. Clear cell RCC overexpressed proximal nephron, angiogenic, and immune response genes, chromophobe RCC oncocytoma overexpressed distal nephron and oxidative phosphorylation genes, papillary RCC overexpressed serine protease inhibitors, and extracellular matrix products, and angiomyolipoma overexpressed muscle developmental, lipid biosynthetic, melanocytic, and distinct angiogenic factors. Quantitative reverse transcriptase-polymerase chain reaction and immunohistochemistry of formalin-fixed renal tumors confirmed overexpression of proximal nephron markers (megalin/low-density lipoprotein-related protein 2, alpha-methylacyl CoA racemase) in clear cell and papillary RCC and distal nephron markers (beta-defensin 1, claudin 7) in chromophobe RCC/oncocytoma. In summary, renal tumor subtypes were classified by distinct gene expression profiles, illustrating tumor pathobiology and translating into novel molecular bioassays using fixed tissue.
Figures

References
-
- Jemal A, Tiwari RC, Murray T, Ghafoor A, Samuels A, Ward E, Feuer EJ, Thun MJ. Cancer Statistics, 2004. CA Cancer J Clin. 2004;54:8–29. - PubMed
-
- Homma Y, Kawabe K, Kitamura T, Nishimura Y, Shinohara M, Kondo Y, Saito I, Minowada S, Asakage Y. Increased incidental detection and reduced mortality in renal cancer—recent retrospective analysis at eight institutions. Int J Urol. 1995;2:77–80. - PubMed
-
- Amin MaB, Amin MiB, Tamboli P, Javidan J, Stricker H, de-Peralta Venturina M, Deshpande A, Menon M. Prognostic impact of histologic subtyping of adult renal epithelial neoplasms: an experience of 405 cases. Am J Surg Pathol. 2002;26:281–291. - PubMed
-
- Zisman A, Pantuck AJ, Wieder J, Chao DH, Dorey F, Said JW, deKernion JB, Figlin RA, Belldegrun AS. Risk group assessment and clinical outcome algorithm to predict the natural history of patients with surgically resected renal cell carcinoma. J Clin Oncol. 2002;20:4559–4566. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

