Recurrent structural RNA motifs, Isostericity Matrices and sequence alignments
- PMID: 15860776
- PMCID: PMC1087784
- DOI: 10.1093/nar/gki535
Recurrent structural RNA motifs, Isostericity Matrices and sequence alignments
Abstract
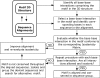
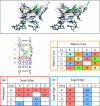
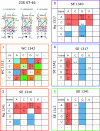
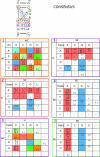
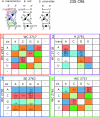
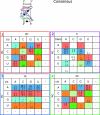
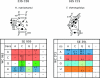
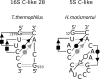
The occurrences of two recurrent motifs in ribosomal RNA sequences, the Kink-turn and the C-loop, are examined in crystal structures and systematically compared with sequence alignments of rRNAs from the three kingdoms of life in order to identify the range of the structural and sequence variations. Isostericity Matrices are used to analyze structurally the sequence variations of the characteristic non-Watson-Crick base pairs for each motif. We show that Isostericity Matrices for non-Watson-Crick base pairs provide important tools for deriving the sequence signatures of recurrent motifs, for scoring and refining sequence alignments, and for determining whether motifs are conserved throughout evolution. The systematic use of Isostericity Matrices identifies the positions of the insertion or deletion of one or more nucleotides relative to the structurally characterized examples of motifs and, most importantly, specifies whether these changes result in new motifs. Thus, comparative analysis coupled with Isostericity Matrices allows one to produce and refine structural sequence alignments. The analysis, based on both sequence and structure, permits therefore the evaluation of the conservation of motifs across phylogeny and the derivation of rules of equivalence between structural motifs. The conservations observed in Isostericity Matrices form a predictive basis for identifying motifs in sequences.
Figures


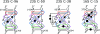
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

