Dissecting eukaryotic translation and its control by ribosome density mapping
- PMID: 15860778
- PMCID: PMC1087779
- DOI: 10.1093/nar/gki331
Dissecting eukaryotic translation and its control by ribosome density mapping
Abstract
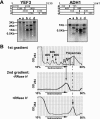
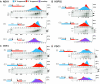
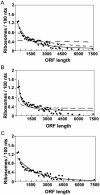
Translation of an mRNA is generally divided into three stages: initiation, elongation and termination. The relative rates of these steps determine both the number and position of ribosomes along the mRNA, but traditional velocity sedimentation assays for the translational status of mRNA determine only the number of bound ribosomes. We developed a procedure, termed Ribosome Density Mapping (RDM), that uses site-specific cleavage of polysomal mRNA followed by separation on a sucrose gradient and northern analysis, to determine the number of ribosomes associated with specified portions of a particular mRNA. This procedure allows us to test models for translation and its control, and to examine properties of individual steps of translation in vivo. We tested specific predictions from the current model for translational control of GCN4 expression in yeast and found that ribosomes were differentially associated with the uORFs elements and coding region under different growth conditions, consistent with this model. We also mapped ribosome density along the ORF of several mRNAs, to probe basic kinetic properties of translational steps in yeast. We found no detectable decline in ribosome density between the 5' and 3' ends of the ORFs, suggesting that the average processivity of elongation is very high. Conversely, there was no queue of ribosomes at the termination site, suggesting that termination is not very slow relative to elongation and initiation. Finally, the RDM results suggest that less frequent initiation of translation on mRNAs with longer ORFs is responsible for the inverse correlation between ORF length and ribosomal density that we observed in a global analysis of translation. These results provide new insights into eukaryotic translation in vivo.
Figures





Similar articles
-
A quantitative model for translational control of the GCN4 gene of Saccharomyces cerevisiae.New Biol. 1991 May;3(5):511-24. New Biol. 1991. PMID: 1883814
-
Post-termination ribosome interactions with the 5'UTR modulate yeast mRNA stability.EMBO J. 1999 Jun 1;18(11):3139-52. doi: 10.1093/emboj/18.11.3139. EMBO J. 1999. PMID: 10357825 Free PMC article.
-
Study of translational control of eukaryotic gene expression using yeast.Ann N Y Acad Sci. 2004 Dec;1038:60-74. doi: 10.1196/annals.1315.012. Ann N Y Acad Sci. 2004. PMID: 15838098
-
Recent mechanistic insights into eukaryotic ribosomes.Curr Opin Cell Biol. 2009 Jun;21(3):435-43. doi: 10.1016/j.ceb.2009.01.023. Epub 2009 Feb 23. Curr Opin Cell Biol. 2009. PMID: 19243929 Review.
-
Detecting ribosomal association with the 5' leader of mRNAs by Ribosome Density Mapping (RDM).Methods Enzymol. 2007;431:163-75. doi: 10.1016/S0076-6879(07)31009-4. Methods Enzymol. 2007. PMID: 17923235 Review.
Cited by
-
A comprehensive, quantitative, and genome-wide model of translation.PLoS Comput Biol. 2010 Jul 29;6(7):e1000865. doi: 10.1371/journal.pcbi.1000865. PLoS Comput Biol. 2010. PMID: 20686685 Free PMC article.
-
Mathematical and Computational Modelling of Ribosomal Movement and Protein Synthesis: an overview.Comput Struct Biotechnol J. 2012 Feb 20;1:e201204002. doi: 10.5936/csbj.201204002. eCollection 2012. Comput Struct Biotechnol J. 2012. PMID: 24688632 Free PMC article. Review.
-
Protein expression regulation under oxidative stress.Mol Cell Proteomics. 2011 Dec;10(12):M111.009217. doi: 10.1074/mcp.M111.009217. Epub 2011 Sep 20. Mol Cell Proteomics. 2011. PMID: 21933953 Free PMC article.
-
Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes.Genome Biol. 2014 Jan 7;15(1):R6. doi: 10.1186/gb-2014-15-1-r6. Genome Biol. 2014. PMID: 24393600 Free PMC article.
-
Mechanistically consistent reduced models of synthetic gene networks.Biophys J. 2013 May 7;104(9):2098-109. doi: 10.1016/j.bpj.2013.03.031. Biophys J. 2013. PMID: 23663853 Free PMC article.
References
-
- Rickwood D. Preparative Centrifugation. A Practical Approach, 2nd edn. Oxford, New York: 1992.
-
- Dickson L.M., Brown A.J. mRNA translation in yeast during entry into stationary phase. Mol. Gen. Genet. 1998;259:282–293. - PubMed
-
- Proweller A., Butler J.S. Ribosomal association of poly(A)-binding protein in poly(A)-deficient Saccharomyces cerevisiae. J. Biol. Chem. 1996;271:10859–10865. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

