The tRNA methylase METTL1 is phosphorylated and inactivated by PKB and RSK in vitro and in cells
- PMID: 15861136
- PMCID: PMC1142581
- DOI: 10.1038/sj.emboj.7600648
The tRNA methylase METTL1 is phosphorylated and inactivated by PKB and RSK in vitro and in cells
Abstract
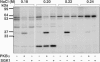
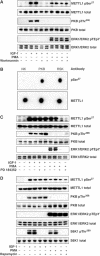
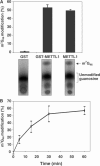
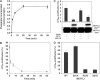
A substrate for protein kinase B (PKB)alpha in HeLa cell extracts was identified as methyltransferase-like protein-1 (METTL1), the orthologue of trm8, which catalyses the 7-methylguanosine modification of tRNA in Saccharomyces cerevisiae. PKB and ribosomal S6 kinase (RSK) both phosphorylated METTL1 at Ser27 in vitro. Ser27 became phosphorylated when HEK293 cells were stimulated with insulin-like growth factor-1 (IGF-1) and this was prevented by inhibition of phosphatidyinositol 3-kinase. The IGF-1-induced Ser27 phosphorylation did not occur in 3-phosphoinositide-dependent protein kinase-1 (PDK1)-deficient embryonic stem cells, but occurred normally in PDK1[L155E] cells, indicating that the effect of IGF-1 is mediated by PKB. METTL1 also became phosphorylated at Ser27 in response to phorbol-12-myristate 13-acetate and this was prevented by PD 184352 or pharmacological inhibition of RSK. Phosphorylation of METTL1 by PKB or RSK inactivated METTL1 in vitro, as did mutation of Ser27 to Asp or Glu. Expression of METTL1[S27D] or METTL1[S27E] did not rescue the growth phenotype of yeast lacking trm8. In contrast, expression of METTL1 or METTL1[S27A] partially rescued growth. These results demonstrate that METTL1 is inactivated by PKB and RSK in cells, and the potential implications of this finding are discussed.
Figures




References
-
- Alessi DR, Caudwell FB, Andjelkovic M, Hemmings BA, Cohen P (1996) Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett 399: 333–338 - PubMed
-
- Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PRJ, Reese CB, Cohen P (1997) Characterisation of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bá. Curr Biol 7: 261–269 - PubMed
-
- Alessi DR, Street AJ, Cohen P, Cohen PW (1993) Inhibitor-2 functions like a chaperone to fold three expressed isoforms of mammalian protein phosphatase-1 into a conformation with the specificity and regulatory properties of the native enzyme. Eur J Biochem 213: 1055–1066 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

