A phase I study of hydralazine to demethylate and reactivate the expression of tumor suppressor genes
- PMID: 15862127
- PMCID: PMC1131894
- DOI: 10.1186/1471-2407-5-44
A phase I study of hydralazine to demethylate and reactivate the expression of tumor suppressor genes
Abstract
Background: The antihypertensive compound hydralazine is a known demethylating agent. This phase I study evaluated the tolerability and its effects upon DNA methylation and gene reactivation in patients with untreated cervical cancer.
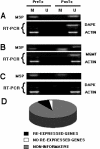
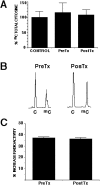
Methods: Hydralazine was administered to cohorts of 4 patients at the following dose levels: I) 50 mg/day, II) 75 mg/day, III) 100 mg/day and IV) 150 mg/day. Tumor biopsies and peripheral blood samples were taken the day before and after treatment. The genes APC, MGMT; ER, GSTP1, DAPK, RARbeta, FHIT and p16 were evaluated pre and post-treatment for DNA promoter methylation and gene expression by MSP (Methylation-Specific PCR) and RT-PCR respectively in each of the tumor samples. Methylation of the imprinted H19 gene and the "normally methylated" sequence clone 1.2 was also analyzed. Global DNA methylation was analyzed by capillary electrophoresis and cytosine extension assay. Toxicity was evaluated using the NCI Common Toxicity Criteria.
Results: Hydralazine was well tolerated. Toxicities were mild being the most common nausea, dizziness, fatigue, headache and palpitations. Overall, 70% of the pretreatment samples and all the patients had at least one methylated gene. Rates of demethylation at the different dose levels were as follows: 50 mg/day, 40%; 75 mg/day, 52%, 100 mg/day, 43%, and 150 mg/day, 32%. Gene expression analysis showed only 12 informative cases, of these 9 (75%) re-expressed the gene. There was neither change in the methylation status of H19 and clone 1.2 nor changes in global DNA methylation.
Conclusion: Hydralazine at doses between 50 and 150 mg/day is well tolerated and effective to demethylate and reactivate the expression of tumor suppressor genes without affecting global DNA methylation.
Figures



References
-
- Beard C, Li E, Jaenisch R. methylation activates Xist in somatic but not in embryonic cells. Genes Dev. 1995;9:2325–34. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

