The nucleocapsid protein of SARS coronavirus has a high binding affinity to the human cellular heterogeneous nuclear ribonucleoprotein A1
- PMID: 15862300
- PMCID: PMC7094256
- DOI: 10.1016/j.febslet.2005.03.080
The nucleocapsid protein of SARS coronavirus has a high binding affinity to the human cellular heterogeneous nuclear ribonucleoprotein A1
Abstract
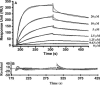
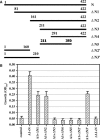
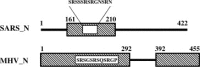
The nucleocapsid (N) protein of SARS coronavirus (SARS_CoV) is a major structural component of virions, which appears to be a multifunctional protein involved in viral RNA replication and translation. Heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) is related to the pre-mRNA splicing in the nucleus and translation regulation in the cytoplasm. In this report, based on the relevant biophysical and biochemical assays, the nucleocapsid protein of SARS_CoV (SARS_N) was discovered to exhibit high binding affinity to human hnRNP A1. GST pull-down results clearly demonstrated that SARS_N protein could directly and specifically bind to human hnRNP A1 in vitro. Yeast two-hybrid assays further indicated in vivo that such binding relates to the fragment (aa 161-210) of SARS_N and the Gly-rich domain (aa 203-320) of hnRNP A1. Moreover, kinetic analyses by surface plasmon resonance (SPR) technology revealed that SARS_N protein has a specific binding affinity against human hnRNP A1 with K(D) at 0.35 +/- 0.02 microM (k(on) = 5.83 +/- 0.42 x 10(3) M(-1)s(-1) and k(off) = 2.06 +/- 0.12 x 10(-3)s(-1)). It is suggested that both SARS_N and hnRNP A1 proteins are possibly within the SARS_CoV replication/transcription complex and SARS_N/human hnRNP A1 interaction might function in the regulation of SARS_CoV RNA synthesis. In addition, the determined results showed that SARS_N protein has only one binding domain for interacting with human hnRNP A1, which is different from the mouse hepatitis virus (MHV) binding case where the nucleocapsid protein of MHV (MHV_N) was found to have two binding domains involved in the MHV_N/hnRNP A1 interaction, thereby suggesting that SARS_N protein might carry out a different binding mode to bind to human hnRNP A1 for its further function performance in comparison with MHV_N.
Figures

Similar articles
-
The nucleocapsid protein of coronavirus mouse hepatitis virus interacts with the cellular heterogeneous nuclear ribonucleoprotein A1 in vitro and in vivo.Virology. 1999 Dec 5;265(1):96-109. doi: 10.1006/viro.1999.0025. Virology. 1999. PMID: 10603321 Free PMC article.
-
Cellular protein hnRNP-A1 interacts with the 3'-end and the intergenic sequence of mouse hepatitis virus negative-strand RNA to form a ribonucleoprotein complex.Adv Exp Med Biol. 1998;440:227-34. doi: 10.1007/978-1-4615-5331-1_28. Adv Exp Med Biol. 1998. PMID: 9782285
-
Multiple type A/B heterogeneous nuclear ribonucleoproteins (hnRNPs) can replace hnRNP A1 in mouse hepatitis virus RNA synthesis.J Virol. 2003 Oct;77(19):10584-93. doi: 10.1128/jvi.77.19.10584-10593.2003. J Virol. 2003. PMID: 12970443 Free PMC article.
-
Heterogeneous nuclear ribonucleoprotein A1 in health and neurodegenerative disease: from structural insights to post-transcriptional regulatory roles.Mol Cell Neurosci. 2013 Sep;56:436-46. doi: 10.1016/j.mcn.2012.12.002. Epub 2012 Dec 14. Mol Cell Neurosci. 2013. PMID: 23247072 Review.
-
A comprehensive understanding of hnRNP A1 role in cancer: new perspectives on binding with noncoding RNA.Cancer Gene Ther. 2023 Mar;30(3):394-403. doi: 10.1038/s41417-022-00571-1. Epub 2022 Dec 2. Cancer Gene Ther. 2023. PMID: 36460805 Review.
Cited by
-
Epidemiology of coronaviruses, genetics, vaccines, and scenario of current pandemic of coronavirus diseases 2019 (COVID-19): a fuzzy set approach.Hum Vaccin Immunother. 2021 May 4;17(5):1296-1303. doi: 10.1080/21645515.2020.1798697. Epub 2021 Mar 15. Hum Vaccin Immunother. 2021. PMID: 33720797 Free PMC article. Review.
-
Characterization of Two Monoclonal Antibodies That Recognize Linker Region and Carboxyl Terminal Domain of Coronavirus Nucleocapsid Protein.PLoS One. 2016 Sep 30;11(9):e0163920. doi: 10.1371/journal.pone.0163920. eCollection 2016. PLoS One. 2016. PMID: 27689694 Free PMC article.
-
Cellular hnRNP A1 Interacts with Nucleocapsid Protein of Porcine Epidemic Diarrhea Virus and Impairs Viral Replication.Viruses. 2018 Mar 13;10(3):127. doi: 10.3390/v10030127. Viruses. 2018. PMID: 29534017 Free PMC article.
-
Membraneless organelles restructured and built by pandemic viruses: HIV-1 and SARS-CoV-2.J Mol Cell Biol. 2021 Aug 4;13(4):259-268. doi: 10.1093/jmcb/mjab020. J Mol Cell Biol. 2021. PMID: 33760045 Free PMC article. Review.
-
SARS-CoV-2 Evolutionary Adaptation toward Host Entry and Recognition of Receptor O-Acetyl Sialylation in Virus-Host Interaction.Int J Mol Sci. 2020 Jun 26;21(12):4549. doi: 10.3390/ijms21124549. Int J Mol Sci. 2020. PMID: 32604730 Free PMC article. Review.
References
-
- Peiris J.S., Chu C.M., Cheng V.C., Chan K.S., Hung I.F., Poon L.L., Law K.I., Tang B.S., Hon T.Y., Chan C.S., Chan K.H., Ng J.S., Zheng B.J., Ng W.L., Lai R.W., Guan Y., Yuen K.Y., Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet, 361, (2003), 1767– 1772. - PMC - PubMed
-
- Drosten C., Gunther S., Preiser W., Van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Osterhaus A.D., Schmitz H., Doerr H.W., Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med., 348, (2003), 1967– 1976. - PubMed
-
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med., 348, (2003), 1953– 1966. - PubMed
-
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L., The Genome sequence of the SARS-associated coronavirus. Science, 300, (2003), 1399– 1404. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

