Constant pH molecular dynamics with proton tautomerism
- PMID: 15863480
- PMCID: PMC1366513
- DOI: 10.1529/biophysj.105.061341
Constant pH molecular dynamics with proton tautomerism
Abstract
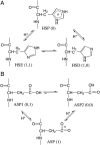
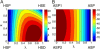
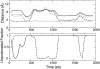
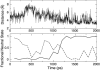
The current article describes a new two-dimensional lambda-dynamics method to include proton tautomerism in continuous constant pH molecular dynamics (CPHMD) simulations. The two-dimensional lambda-dynamics framework is used to devise a tautomeric state titration model for the CPHMD simulations involving carboxyl and histidine residues. Combined with the GBSW implicit solvent model, the new method is tested on titration simulations of blocked histidine and aspartic acid as well as two benchmark proteins, turkey ovomucoid third domain (OMTKY3) and ribonuclease A (RNase A). A detailed analysis of the errors inherent to the CPHMD methodology as well as those due to the underlying solvation model is given. The average absolute error for the computed pKa values in OMTKY3 is 1.0 pK unit. In RNase A the average absolute errors for the carboxyl and histidine residues are 1.6 and 0.6 pK units, respectively. In contrast to the previous work, the new model predicts the correct sign for all the pKa shifts, but one, in the benchmark proteins. The predictions of the tautomeric states of His12 and His48 and the conformational states of His48 and His119 are in agreement with experiment. Based on the simulations of OMTKY3 and RNase A, the current work has demonstrated the capability of the CPHMD technique in revealing pH-coupled conformational dynamics of protein side chains.
Figures

Similar articles
-
GPU-Accelerated Implementation of Continuous Constant pH Molecular Dynamics in Amber: pKa Predictions with Single-pH Simulations.J Chem Inf Model. 2019 Nov 25;59(11):4821-4832. doi: 10.1021/acs.jcim.9b00754. Epub 2019 Nov 14. J Chem Inf Model. 2019. PMID: 31661616 Free PMC article.
-
Toward accurate prediction of pKa values for internal protein residues: the importance of conformational relaxation and desolvation energy.Proteins. 2011 Dec;79(12):3364-73. doi: 10.1002/prot.23080. Epub 2011 Jul 11. Proteins. 2011. PMID: 21748801
-
Two conformational states of Turkey ovomucoid third domain at low pH: three-dimensional structures, internal dynamics, and interconversion kinetics and thermodynamics.Biochemistry. 2003 Jun 3;42(21):6380-91. doi: 10.1021/bi034053f. Biochemistry. 2003. PMID: 12767219
-
Biomolecular simulations at constant pH.Curr Opin Struct Biol. 2005 Apr;15(2):157-63. doi: 10.1016/j.sbi.2005.02.002. Curr Opin Struct Biol. 2005. PMID: 15837173 Review.
-
Advances in implicit models of water solvent to compute conformational free energy and molecular dynamics of proteins at constant pH.Adv Protein Chem Struct Biol. 2011;85:281-322. doi: 10.1016/B978-0-12-386485-7.00008-9. Adv Protein Chem Struct Biol. 2011. PMID: 21920327 Review.
Cited by
-
Thermodynamic coupling of protonation and conformational equilibria in proteins: theory and simulation.Biophys J. 2012 Apr 4;102(7):1590-7. doi: 10.1016/j.bpj.2012.02.021. Epub 2012 Apr 3. Biophys J. 2012. PMID: 22500759 Free PMC article.
-
phbuilder: A Tool for Efficiently Setting up Constant pH Molecular Dynamics Simulations in GROMACS.J Chem Inf Model. 2024 Feb 12;64(3):567-574. doi: 10.1021/acs.jcim.3c01313. Epub 2024 Jan 12. J Chem Inf Model. 2024. PMID: 38215282 Free PMC article.
-
Role of Pseudoisocytidine Tautomerization in Triplex-Forming Oligonucleotides: In Silico and in Vitro Studies.ACS Omega. 2017 May 31;2(5):2165-2177. doi: 10.1021/acsomega.7b00347. Epub 2017 May 17. ACS Omega. 2017. PMID: 30023656 Free PMC article.
-
GPU-Accelerated Implementation of Continuous Constant pH Molecular Dynamics in Amber: pKa Predictions with Single-pH Simulations.J Chem Inf Model. 2019 Nov 25;59(11):4821-4832. doi: 10.1021/acs.jcim.9b00754. Epub 2019 Nov 14. J Chem Inf Model. 2019. PMID: 31661616 Free PMC article.
-
Accurately Predicting Protein pKa Values Using Nonequilibrium Alchemy.J Chem Theory Comput. 2023 Nov 14;19(21):7833-7845. doi: 10.1021/acs.jctc.3c00721. Epub 2023 Oct 11. J Chem Theory Comput. 2023. PMID: 37820376 Free PMC article.
References
-
- Matthew, J. B., F. R. Gurd, B. Garcia-Moreno, M. A. Flanagan, K. L. March, and S. J. Shire. 1985. pH-dependent processes in proteins. CRC Crit. Rev. Biochem. 18:91–197. - PubMed
-
- Kelly, J. W. 1997. Amyloid fibril formation and protein misassembly: a structural quest for insights into amyloid and prion diseases. Structure. 5:595–600. - PubMed
-
- Clippingdale, A. B., J. D. Wade, and C. J. Barrow. 2001. The amyloid-β peptide and its role in Alzheimer's disease. J. Pept. Sci. 7:227–249. - PubMed
-
- Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of Influenza haemagglutinin at the pH of membrane fusion. Nature. 371:37–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

