Chevron behavior and isostable enthalpic barriers in protein folding: successes and limitations of simple Gō-like modeling
- PMID: 15863486
- PMCID: PMC1366551
- DOI: 10.1529/biophysj.104.057471
Chevron behavior and isostable enthalpic barriers in protein folding: successes and limitations of simple Gō-like modeling
Abstract
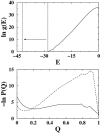
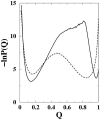
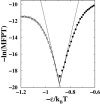
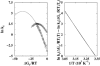
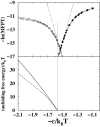
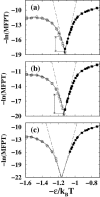
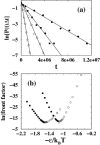
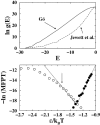
It has been demonstrated that a "near-Levinthal" cooperative mechanism, whereby the common Gō interaction scheme is augmented by an extra favorability for the native state as a whole, can lead to apparent two-state folding/unfolding kinetics over a broad range of native stabilities in lattice models of proteins. Here such a mechanism is shown to be generalizable to a simplified continuum (off-lattice) Langevin dynamics model with a Calpha protein chain representation, with the resulting chevron plots exhibiting an extended quasilinear regime reminiscent of that of apparent two-state real proteins. Similarly high degrees of cooperativity are possible in Gō-like continuum models with rudimentary pairwise desolvation barriers as well. In these models, cooperativity increases with increasing desolvation barrier height, suggesting strongly that two-state-like folding/unfolding kinetics would be achievable when the pairwise desolvation barrier becomes sufficiently high. Besides cooperativity, another generic folding property of interest that has emerged from published experiments on several apparent two-state proteins is that their folding relaxation under constant native stability (isostability) conditions is essentially Arrhenius, entailing high intrinsic enthalpic folding barriers of approximately 17-30 kcal/mol. Based on a new analysis of published data on barnase, here we propose that a similar property should also apply to a certain class of non-two-state proteins that fold with chevron rollovers. However, several continuum Gō-like constructs considered here fail to predict any significant intrinsic enthalpic folding barrier under isostability conditions; thus the physical origin of such barriers in real proteins remains to be elucidated.
Figures






Similar articles
-
Solvation and desolvation effects in protein folding: native flexibility, kinetic cooperativity and enthalpic barriers under isostability conditions.Phys Biol. 2005 Nov 9;2(4):S75-85. doi: 10.1088/1478-3975/2/4/S01. Phys Biol. 2005. PMID: 16280624
-
Desolvation is a likely origin of robust enthalpic barriers to protein folding.J Mol Biol. 2005 Jun 17;349(4):872-89. doi: 10.1016/j.jmb.2005.03.084. Epub 2005 Apr 15. J Mol Biol. 2005. PMID: 15893325
-
Simple two-state protein folding kinetics requires near-levinthal thermodynamic cooperativity.Proteins. 2003 Sep 1;52(4):510-23. doi: 10.1002/prot.10506. Proteins. 2003. PMID: 12910451
-
Cooperativity, local-nonlocal coupling, and nonnative interactions: principles of protein folding from coarse-grained models.Annu Rev Phys Chem. 2011;62:301-26. doi: 10.1146/annurev-physchem-032210-103405. Annu Rev Phys Chem. 2011. PMID: 21453060 Review.
-
Protein folding in the landscape perspective: chevron plots and non-Arrhenius kinetics.Proteins. 1998 Jan;30(1):2-33. doi: 10.1002/(sici)1097-0134(19980101)30:1<2::aid-prot2>3.0.co;2-r. Proteins. 1998. PMID: 9443337 Review.
Cited by
-
Discrete kinetic models from funneled energy landscape simulations.PLoS One. 2012;7(12):e50635. doi: 10.1371/journal.pone.0050635. Epub 2012 Dec 12. PLoS One. 2012. PMID: 23251375 Free PMC article.
-
Excluded volume, local structural cooperativity, and the polymer physics of protein folding rates.Proc Natl Acad Sci U S A. 2007 Jun 26;104(26):10841-6. doi: 10.1073/pnas.0609321104. Epub 2007 Jun 14. Proc Natl Acad Sci U S A. 2007. PMID: 17569785 Free PMC article.
-
Distinguishing between cooperative and unimodal downhill protein folding.Proc Natl Acad Sci U S A. 2007 Jan 2;104(1):123-7. doi: 10.1073/pnas.0609717104. Proc Natl Acad Sci U S A. 2007. PMID: 17200301 Free PMC article.
-
Enthalpic barriers dominate the folding and unfolding of the human Cu, Zn superoxide dismutase monomer.J Mol Biol. 2012 Dec 7;424(3-4):192-202. doi: 10.1016/j.jmb.2012.09.009. Epub 2012 Sep 18. J Mol Biol. 2012. PMID: 22999954 Free PMC article.
-
Cooperativity and the origins of rapid, single-exponential kinetics in protein folding.Protein Sci. 2006 Jul;15(7):1608-18. doi: 10.1110/ps.062180806. Protein Sci. 2006. PMID: 16815915 Free PMC article.
References
-
- Jackson, S. E., and A. R. Fersht. 1991. Folding of chymotrypsin inhibitor 2. I. Evidence for a two-state transition. Biochemistry. 30:10428–10435. - PubMed
-
- Chan, H. S., S. Shimizu, and H. Kaya. 2004. Cooperativity principles in protein folding. Methods Enzymol. 380:350–379. - PubMed
-
- Kuhlman, B., D. L. Luisi, P. A. Evans, and D. P. Raleigh. 1998. Global analysis of the effects of temperature and denaturant on the folding and unfolding kinetics of the N-terminal domain of the protein L9. J. Mol. Biol. 284:1661–1670. - PubMed
-
- Daggett, V. 2002. Molecular dynamics simulations of the protein unfolding/folding reaction. Acc. Chem. Res. 35:422–429. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

