Discovery of a unique Ig heavy-chain isotype (IgT) in rainbow trout: Implications for a distinctive B cell developmental pathway in teleost fish
- PMID: 15863615
- PMCID: PMC1100771
- DOI: 10.1073/pnas.0500027102
Discovery of a unique Ig heavy-chain isotype (IgT) in rainbow trout: Implications for a distinctive B cell developmental pathway in teleost fish
Abstract
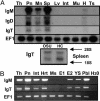
During the analysis of Ig superfamily members within the available rainbow trout (Oncorhynchus mykiss) EST gene index, we identified a unique Ig heavy-chain (IgH) isotype. cDNAs encoding this isotype are composed of a typical IgH leader sequence and a VDJ rearranged segment followed by four Ig superfamily C-1 domains represented as either membrane-bound or secretory versions. Because teleost fish were previously thought to encode and express only two IgH isotypes (IgM and IgD) for their humoral immune repertoire, we isolated all three cDNA isotypes from a single homozygous trout (OSU-142) to confirm that all three are indeed independent isotypes. Bioinformatic and phylogenetic analysis indicates that this previously undescribed divergent isotype is restricted to bony fish, thus we have named this isotype "IgT" (tau) for teleost fish. Genomic sequence analysis of an OSU-142 bacterial artificial chromosome (BAC) clone positive for all three IgH isotypes revealed that IgT utilizes the standard rainbow trout V(H) families, but surprisingly, the IgT isotype possesses its own exclusive set of D(H) and J(H) elements for the generation of diversity. The IgT D and J segments and tau constant (C) region genes are located upstream of the D and J elements for IgM, representing a genomic IgH architecture that has not been observed in any other vertebrate class. All three isotypes are primarily expressed in the spleen and pronephros (bone marrow equivalent), and ontogenically, expression of IgT is present 4 d before hatching in developing embryos.
Figures


Similar articles
-
Discovery of a new class of immunoglobulin heavy chain from fugu.Eur J Immunol. 2005 Nov;35(11):3320-31. doi: 10.1002/eji.200535248. Eur J Immunol. 2005. PMID: 16224815
-
Identification and characterization of a novel immunoglobulin Z isotype in zebrafish: implications for a distinct B cell receptor in lower vertebrates.Mol Immunol. 2010 Jan;47(4):738-46. doi: 10.1016/j.molimm.2009.10.010. Epub 2009 Nov 20. Mol Immunol. 2010. PMID: 19931913
-
Identification, characterization and genetic mapping of TLR7, TLR8a1 and TLR8a2 genes in rainbow trout (Oncorhynchus mykiss).Dev Comp Immunol. 2010 Feb;34(2):219-33. doi: 10.1016/j.dci.2009.10.002. Epub 2009 Oct 23. Dev Comp Immunol. 2010. PMID: 19825389
-
Antibody repertoire development in swine.Dev Comp Immunol. 2006;30(1-2):199-221. doi: 10.1016/j.dci.2005.06.025. Dev Comp Immunol. 2006. PMID: 16168480 Review.
-
Immunoglobulins in teleosts.Immunogenetics. 2021 Feb;73(1):65-77. doi: 10.1007/s00251-020-01195-1. Epub 2021 Jan 13. Immunogenetics. 2021. PMID: 33439286 Review.
Cited by
-
The pyloric caeca area is a major site for IgM(+) and IgT(+) B cell recruitment in response to oral vaccination in rainbow trout.PLoS One. 2013 Jun 13;8(6):e66118. doi: 10.1371/journal.pone.0066118. Print 2013. PLoS One. 2013. PMID: 23785475 Free PMC article.
-
Systematic characterization of immunoglobulin loci and deep sequencing of the expressed repertoire in the Atlantic cod (Gadus morhua).BMC Genomics. 2024 Jul 3;25(1):663. doi: 10.1186/s12864-024-10571-0. BMC Genomics. 2024. PMID: 38961347 Free PMC article.
-
Differential expression of innate and adaptive immune genes in the survivors of three gibel carp gynogenetic clones after herpesvirus challenge.BMC Genomics. 2019 May 28;20(1):432. doi: 10.1186/s12864-019-5777-z. BMC Genomics. 2019. PMID: 31138127 Free PMC article.
-
Fish Immunoglobulins.Biology (Basel). 2016 Nov 21;5(4):45. doi: 10.3390/biology5040045. Biology (Basel). 2016. PMID: 27879632 Free PMC article. Review.
-
Dissecting teleost B cell differentiation using transcription factors.Dev Comp Immunol. 2011 Sep;35(9):898-905. doi: 10.1016/j.dci.2011.01.009. Epub 2011 Jan 17. Dev Comp Immunol. 2011. PMID: 21251922 Free PMC article. Review.
References
-
- Eason, D. D., Cannon, J. P., Haire, R. N., Rast, J. P., Ostrov, D. A. & Litman, G. W. (2004) Semin. Immunol. 16, 215–226. - PubMed
-
- Flajnik, M. F. (2002) Nat. Rev. Immunol. 2, 688–698. - PubMed
-
- Hordvik, I., Thevarajan, J., Samdal, I., Bastani, N. & Krossoy, B. (1999) Scand. J. Immunol. 50, 202–210. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

