The Arabidopsis IspH homolog is involved in the plastid nonmevalonate pathway of isoprenoid biosynthesis
- PMID: 15863698
- PMCID: PMC1150385
- DOI: 10.1104/pp.104.058735
The Arabidopsis IspH homolog is involved in the plastid nonmevalonate pathway of isoprenoid biosynthesis
Abstract
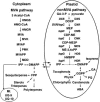
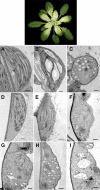
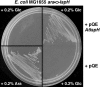
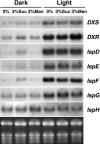
Plant isoprenoids are synthesized via two independent pathways, the cytosolic mevalonate (MVA) pathway and the plastid nonmevalonate pathway. The Escherichia coli IspH (LytB) protein is involved in the last step of the nonmevalonate pathway. We have isolated an Arabidopsis (Arabidopsis thaliana) ispH null mutant that has an albino phenotype and have generated Arabidopsis transgenic lines showing various albino patterns caused by IspH transgene-induced gene silencing. The initiation of albino phenotypes rendered by IspH gene silencing can arise independently from multiple sites of the same plant. After a spontaneous initiation, the albino phenotype is systemically spread toward younger tissues along the source-to-sink flow relative to the initiation site. The development of chloroplasts is severely impaired in the IspH-deficient albino tissues. Instead of thylakoids, mutant chloroplasts are filled with vesicles. Immunoblot analysis reveals that Arabidopsis IspH is a chloroplast stromal protein. Expression of Arabidopsis IspH complements the lethal phenotype of an E. coli ispH mutant. In 2-week-old Arabidopsis seedlings, the expression of 1-deoxy-d-xylulose 5-phosphate synthase (DXS), 1-deoxy-d-xylulose 5-phosphate reductoisomerase (DXR), IspD, IspE, IspF, and IspG genes is induced by light, whereas the expression of the IspH gene is constitutive. The addition of 3% sucrose in the media slightly increased levels of DXS, DXR, IspD, IspE, and IspF mRNA in the dark. In a 16-h-light/8-h-dark photoperiod, the accumulation of the IspH transcript oscillates with the highest levels detected in the early light period (2-6 h) and the late dark period (4-6 h). The expression patterns of DXS and IspG are similar to that of IspH, indicating that these genes are coordinately regulated in Arabidopsis when grown in a 16-h-light/8-h-dark photoperiod.
Figures





Similar articles
-
Functional evidence for the involvement of Arabidopsis IspF homolog in the nonmevalonate pathway of plastid isoprenoid biosynthesis.Planta. 2006 Mar;223(4):779-84. doi: 10.1007/s00425-005-0140-9. Epub 2005 Oct 18. Planta. 2006. PMID: 16231155
-
Functional analysis of the final steps of the 1-deoxy-D-xylulose 5-phosphate (DXP) pathway to isoprenoids in plants using virus-induced gene silencing.Plant Physiol. 2004 Apr;134(4):1401-13. doi: 10.1104/pp.103.038133. Epub 2004 Apr 2. Plant Physiol. 2004. PMID: 15064370 Free PMC article.
-
Chloroplast localization of methylerythritol 4-phosphate pathway enzymes and regulation of mitochondrial genes in ispD and ispE albino mutants in Arabidopsis.Plant Mol Biol. 2008 Apr;66(6):663-73. doi: 10.1007/s11103-008-9297-5. Epub 2008 Jan 31. Plant Mol Biol. 2008. PMID: 18236010
-
Bioorganometallic chemistry with IspG and IspH: structure, function, and inhibition of the [Fe(4)S(4)] proteins involved in isoprenoid biosynthesis.Angew Chem Int Ed Engl. 2014 Apr 22;53(17):4294-310. doi: 10.1002/anie.201306712. Epub 2014 Jan 31. Angew Chem Int Ed Engl. 2014. PMID: 24481599 Free PMC article. Review.
-
Functional Gene Network of Prenyltransferases in Arabidopsis thaliana.Molecules. 2019 Dec 12;24(24):4556. doi: 10.3390/molecules24244556. Molecules. 2019. PMID: 31842481 Free PMC article. Review.
Cited by
-
2C-Methyl- D- erythritol 2,4-cyclodiphosphate synthase from Stevia rebaudiana Bertoni is a functional gene.Mol Biol Rep. 2012 Dec;39(12):10971-8. doi: 10.1007/s11033-012-1998-9. Epub 2012 Oct 12. Mol Biol Rep. 2012. PMID: 23065206
-
Functional characterization and evolution of the isotuberculosinol operon in Mycobacterium tuberculosis and related Mycobacteria.Front Microbiol. 2012 Oct 12;3:368. doi: 10.3389/fmicb.2012.00368. eCollection 2012. Front Microbiol. 2012. PMID: 23091471 Free PMC article.
-
Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes.Genome Biol. 2006;7(8):R76. doi: 10.1186/gb-2006-7-8-R76. Epub 2006 Aug 17. Genome Biol. 2006. PMID: 16916443 Free PMC article.
-
Editing of accD and ndhF chloroplast transcripts is partially affected in the Arabidopsis vanilla cream1 mutant.Plant Mol Biol. 2010 Jun;73(3):309-23. doi: 10.1007/s11103-010-9616-5. Epub 2010 Feb 9. Plant Mol Biol. 2010. PMID: 20143129
-
Disruption of the homogentisate solanesyltransferase gene results in albino and dwarf phenotypes and root, trichome and stomata defects in Arabidopsis thaliana.PLoS One. 2014 Apr 17;9(4):e94031. doi: 10.1371/journal.pone.0094031. eCollection 2014. PLoS One. 2014. PMID: 24743244 Free PMC article.
References
-
- Altincicek B, Duin EC, Reichenberg A, Hedderich R, Kollas AK, Hintz M, Wagner S, Wiesner J, Beck E, Jomaa H (2002) LytB protein catalyzes the terminal step of the 2-C-methyl-d-erythritol-4-phosphate pathway of isoprenoid biosynthesis. FEBS Lett 532: 437–440 - PubMed
-
- Altincicek B, Kollas A, Eberl M, Wiesner J, Sanderbrand S, Hintz M, Beck F, Jomaa H (2001) LytB, a novel gene of the 2-C-methyl-d-erythritol 4-phosphate pathway of isoprenoid biosynthesis in Escherichia coli. FEBS Lett 499: 37–40 - PubMed
-
- Bick JA, Lange BM (2003) Metabolic cross talk between cytosolic and plastidial pathways of isoprenoid biosynthesis: unidirectional transport of intermediates across the chloroplast envelope membrane. Arch Biochem Biophys 415: 146–154 - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

