Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis
- PMID: 15863702
- PMCID: PMC1150392
- DOI: 10.1104/pp.104.053884
Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis
Abstract
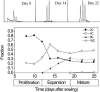
Organ growth results from the progression of component cells through subsequent phases of proliferation and expansion before reaching maturity. We combined kinematic analysis, flowcytometry, and microarray analysis to characterize cell cycle regulation during the growth process of leaves 1 and 2 of Arabidopsis (Arabidopsis thaliana). Kinematic analysis showed that the epidermis proliferates until day 12; thereafter, cells expand until day 19 when leaves reach maturity. Flowcytometry revealed that endoreduplication occurs from the time cell division rates decline until the end of cell expansion. Analysis of 10 time points with a 6k-cDNA microarray showed that transitions between the growth stages were closely reflected in the mRNA expression data. Subsequent genome-wide microarray analysis on the three main stages allowed us to categorize known cell cycle genes into three major classes: constitutively expressed, proliferative, and inhibitory. Comparison with published expression data obtained from root zones corresponding to similar developmental stages and from synchronized cell cultures supported this categorization and enabled us to identify a high confidence set of 131 proliferation genes. Most of those had an M phase-dependent expression pattern and, in addition to many known cell cycle-related genes, there were at least 90 that were unknown or previously not associated with proliferation.
Figures




References
-
- Beemster GTS, Fiorani F, Inzé D (2003) Cell cycle: the key to plant growth control? Trends Plant Sci 8: 154–158 - PubMed
-
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 - PubMed
-
- Boudolf V, Vlieghe K, Beemster GTS, Magyar Z, Torres-Acosta JA, Maes S, Van Der Schueren E, Inze D, De Veylder L (2004) The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16: 2683–2692 - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

