Resurrecting ancestral alcohol dehydrogenases from yeast
- PMID: 15864308
- PMCID: PMC3618678
- DOI: 10.1038/ng1553
Resurrecting ancestral alcohol dehydrogenases from yeast
Abstract
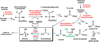
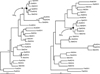
Modern yeast living in fleshy fruits rapidly convert sugars into bulk ethanol through pyruvate. Pyruvate loses carbon dioxide to produce acetaldehyde, which is reduced by alcohol dehydrogenase 1 (Adh1) to ethanol, which accumulates. Yeast later consumes the accumulated ethanol, exploiting Adh2, an Adh1 homolog differing by 24 (of 348) amino acids. As many microorganisms cannot grow in ethanol, accumulated ethanol may help yeast defend resources in the fruit. We report here the resurrection of the last common ancestor of Adh1 and Adh2, called Adh(A). The kinetic behavior of Adh(A) suggests that the ancestor was optimized to make (not consume) ethanol. This is consistent with the hypothesis that before the Adh1-Adh2 duplication, yeast did not accumulate ethanol for later consumption but rather used Adh(A) to recycle NADH generated in the glycolytic pathway. Silent nucleotide dating suggests that the Adh1-Adh2 duplication occurred near the time of duplication of several other proteins involved in the accumulation of ethanol, possibly in the Cretaceous age when fleshy fruits arose. These results help to connect the chemical behavior of these enzymes through systems analysis to a time of global ecosystem change, a small but useful step towards a planetary systems biology.
Figures
Comment in
-
The gene duplication that greased society's wheels.Nat Genet. 2005 Jun;37(6):566-7. doi: 10.1038/ng0605-566. Nat Genet. 2005. PMID: 15920515 No abstract available.
References
-
- Pretorius IS. Tailoring wine yeasts for the new millennium: novel approaches to the ancient art of winemaking. Yeast. 2000;16:675–729. - PubMed
-
- Thornton JW. Resurrecting ancient genes. Experimental analysis of extinct molecules. Nat. Rev. Genet. 2004;5:366–376. - PubMed
-
- Boulton B, Singleton VL, Bisson LF, Kunkee RE. Principles and Practices of Winemaking. New York: Chapman and Hall; 1996. Yeast and biochemistry of ethanol fermentation; pp. 139–172.
-
- Fleet GH, Heard GM. Wine Microbiology and Biotechnology. Chur, Switzerland: Harwood Academic Publishers; 1993. Yeast growth during fermentation; pp. 27–54.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

