T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer
- PMID: 15867090
- PMCID: PMC2213187
- DOI: 10.1084/jem.20050572
T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer
Abstract
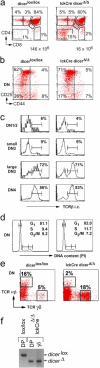
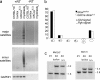
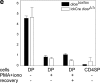
The ribonuclease III enzyme Dicer is essential for the processing of micro-RNAs (miRNAs) and small interfering RNAs (siRNAs) from double-stranded RNA precursors. miRNAs and siRNAs regulate chromatin structure, gene transcription, mRNA stability, and translation in a wide range of organisms. To provide a model system to explore the role of Dicer-generated RNAs in the differentiation of mammalian cells in vivo, we have generated a conditional Dicer allele. Deletion of Dicer at an early stage of T cell development compromised the survival of alphabeta lineage cells, whereas the numbers of gammadelta-expressing thymocytes were not affected. In developing thymocytes, Dicer was not required for the maintenance of transcriptional silencing at pericentromeric satellite sequences (constitutive heterochromatin), the maintenance of DNA methylation and X chromosome inactivation in female cells (facultative heterochromatin), and the stable shutdown of a developmentally regulated gene (developmentally regulated gene silencing). Most remarkably, given that one third of mammalian mRNAs are putative miRNA targets, Dicer seems to be dispensable for CD4/8 lineage commitment, a process in which epigenetic regulation of lineage choice has been well documented. Thus, although Dicer seems to be critical for the development of the early embryo, it may have limited impact on the implementation of some lineage-specific gene expression programs.
Figures



References
-
- Volpe, T.A., C. Kidner, I.M. Hall, G. Teng, S.I. Grewal, and R.A. Martienssen. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 297:1833–1837. - PubMed
-
- Bartel, D.P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116:281–297. - PubMed
-
- Kawasaki, H., and K. Taira. 2004. Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature. 431:211–217. - PubMed
-
- Morris, K.V., S.W. Chan, S.E. Jacobsen, and D.J. Looney. 2004. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 305:1289–1292. - PubMed
-
- Fukagawa, T., M. Nogami, M. Yoshikawa, M. Ikeno, T. Okazaki, Y. Takami, T. Nakayama, and M. Oshimura. 2004. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 6:784–791. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

