Engineering of protease variants exhibiting high catalytic activity and exquisite substrate selectivity
- PMID: 15867160
- PMCID: PMC1100772
- DOI: 10.1073/pnas.0500063102
Engineering of protease variants exhibiting high catalytic activity and exquisite substrate selectivity
Abstract
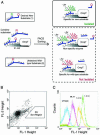
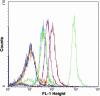
The exquisite selectivity and catalytic activity of enzymes have been shaped by the effects of positive and negative selection pressure during the course of evolution. In contrast, enzyme variants engineered by using in vitro screening techniques to accept novel substrates typically display a higher degree of catalytic promiscuity and lower total turnover in comparison with their natural counterparts. Using bacterial display and multiparameter flow cytometry, we have developed a novel methodology for emulating positive and negative selective pressure in vitro for the isolation of enzyme variants with reactivity for desired novel substrates, while simultaneously excluding those with reactivity toward undesired substrates. Screening of a large library of random mutants of the Escherichia coli endopeptidase OmpT led to the isolation of an enzyme variant, 1.3.19, that cleaved an Ala-Arg peptide bond instead of the Arg-Arg bond preferred by the WT enzyme. Variant 1.3.19 exhibited greater than three million-fold selectivity (-Ala-Arg-/-Arg-Arg-) and a catalytic efficiency for Ala-Arg cleavage that is the same as that displayed by the parent for the preferred substrate, Arg-Arg. A single amino acid Ser223Arg substitution was shown to recapitulate completely the unique catalytic properties of the 1.3.19 variant. These results can be explained by proposing that this mutation acts to "swap" the P(1) Arg side chain normally found in WT substrate peptides with the 223Arg side chain in the S(1) subsite of OmpT.
Figures
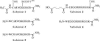

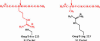
References
-
- Carter, P. & Wells, J. A. (1987) Science 237, 394–399. - PubMed
-
- Bone, R., Fujishige, A., Kettner, C. A. & Agard, D. D. (1991) Biochemistry 30, 10388–10398. - PubMed
-
- Graham, L. A., Brandt, U., Sargent, J. S. & Trumpower, B. L. (1993) J. Bioenerg. Biomembr. 25, 245–257. - PubMed
-
- Harris, J. L. & Craik, C. S. (1998) Curr. Opin. Chem. Biol. 2, 127–132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

