Under the genomic radar: the stealth model of Alu amplification
- PMID: 15867427
- PMCID: PMC1088293
- DOI: 10.1101/gr.3492605
Under the genomic radar: the stealth model of Alu amplification
Abstract
Alu elements are the most successful SINEs (Short INterspersed Elements) in primate genomes and have reached more than 1,000,000 copies in the human genome. The amplification of most Alu elements is thought to occur through a limited number of hyperactive "master" genes that produce a high number of copies during long evolutionary periods of time. However, the existence of long-lived, low-activity Alu lineages in the human genome suggests a more complex propagation mechanism. Using both computational and wet-bench approaches, we reconstructed the evolutionary history of the AluYb lineage, one of the most active Alu lineages in the human genome. We show that the major AluYb lineage expansion in humans is a species-specific event, as nonhuman primates possess only a handful of AluYb elements. However, the oldest existing AluYb element resided in an orthologous position in all hominoid primate genomes examined, demonstrating that the AluYb lineage originated 18-25 million years ago. Thus, the history of the AluYb lineage is characterized by approximately 20 million years of retrotranspositional quiescence preceding a major expansion in the human genome within the past few million years. We suggest that the evolutionary success of the Alu family may be driven at least in part by "stealth-driver" elements that maintain low retrotranspositional activity over extended periods of time and occasionally produce short-lived hyperactive copies responsible for the formation and remarkable expansion of Alu elements within the genome.
Figures

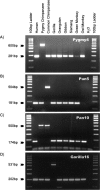
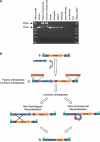

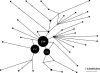

References
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. - PubMed
-
- Bandelt, H.J., Forster, P., and Rohl, A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16: 37–48. - PubMed
-
- Batzer, M.A. and Deininger, P.L. 2002. Alu repeats and human genomic diversity. Nat. Rev. Genet. 3: 370–379. - PubMed
-
- Batzer, M.A., Rubin, C.M., Hellmann-Blumberg, U., Alegria-Hartman, M., Leeflang, E.P., Stern, J.D., Bazan, H.A., Shaikh, T.H., Deininger, P.L., and Schmid, C.W. 1995. Dispersion and insertion polymorphism in two small subfamilies of recently amplified human Alu repeats. J. Mol. Biol. 247: 418–427. - PubMed
-
- Batzer, M.A., Deininger, P.L., Hellmann-Blumberg, U., Jurka, J., Labuda, D., Rubin, C.M., Schmid, C.W., Zietkiewicz, E., and Zuckerkandl, E. 1996. Standardized nomenclature for Alu repeats. J. Mol. Evol. 42: 3–6. - PubMed
Web site references
-
- http://batzerlab.lsu.edu; Batzer laboratory.
-
- http://www.ensembl.org/multi/blastview; Basic Local Alignment Search Tool (BLAST). - PubMed
-
- http://www.fluxus-engineering.com/sharenet.htm; NETWORK 4.1.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
