Structural and sequence motifs of protein (histone) methylation enzymes
- PMID: 15869391
- PMCID: PMC2733851
- DOI: 10.1146/annurev.biophys.34.040204.144452
Structural and sequence motifs of protein (histone) methylation enzymes
Abstract
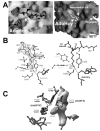
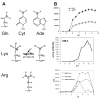
With genome sequencing nearing completion for the model organisms used in biomedical research, there is a rapidly growing appreciation that proteomics, the study of covalent modification to proteins, and transcriptional regulation will likely dominate the research headlines in the next decade. Protein methylation plays a central role in both of these fields, as several different residues (Arg, Lys, Gln) are methylated in cells and methylation plays a central role in the "histone code" that regulates chromatin structure and impacts transcription. In some cases, a single lysine can be mono-, di-, or trimethylated, with different functional consequences for each of the three forms. This review describes structural aspects of methylation of histone lysine residues by two enzyme families with entirely different structural scaffolding (the SET proteins and Dot1p) and methylation of protein arginine residues by PRMTs.
Figures






References
-
- An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–48. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
