CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1
- PMID: 15870271
- PMCID: PMC1087704
- DOI: 10.1128/MCB.25.10.3967-3981.2005
CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1
Abstract
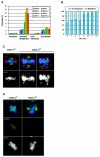
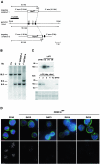
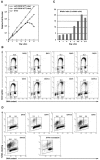
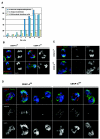
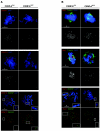
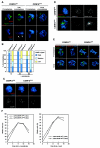
CENP-A is an evolutionarily conserved, centromere-specific variant of histone H3 that is thought to play a central role in directing kinetochore assembly and in centromere function. Here, we have analyzed the consequences of disrupting the CENP-A gene in the chicken DT40 cell line. In CENP-A-depleted cells, kinetochore protein assembly is impaired, as indicated by mislocalization of the inner kinetochore proteins CENP-I, CENP-H, and CENP-C as well as the outer components Nuf2/Hec1, Mad2, and CENP-E. However, BubR1 and the inner centromere protein INCENP are efficiently recruited to kinetochores. Following CENP-A depletion, chromosomes are deficient in proper congression on the mitotic spindle and there is a transient delay in prometaphase. CENP-A-depleted cells further proceed through anaphase and cytokinesis with unequal chromosome segregation, suggesting that some kinetochore function remains following substantial depletion of CENP-A. We furthermore demonstrate that CENP-A-depleted cells exhibit a specific defect in maintaining kinetochore localization of the checkpoint protein BubR1 under conditions of checkpoint activation. Our data thus point to a specific role for CENP-A in assembly of kinetochores competent in the maintenance of mitotic checkpoint signaling.
Figures
References
-
- Adams, R. R., M. Carmena, and W. C. Earnshaw. 2001. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 11:49-54. - PubMed
-
- Biggins, S., and C. E. Walczak. 2003. Captivating capture: how microtubules attach to kinetochores. Curr. Biol. 13:R449-R460. - PubMed
-
- Black, B. E., D. R. Foltz, S. Chakravarthy, K. Luger, V. L. Woods, Jr., and D. W. Cleveland. 2004. Structural determinants for generating centromeric chromatin. Nature 430:578-582. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
