Expression, regulation, and requirement of the toll transmembrane protein during dorsal vessel formation in Drosophila melanogaster
- PMID: 15870289
- PMCID: PMC1087703
- DOI: 10.1128/MCB.25.10.4200-4210.2005
Expression, regulation, and requirement of the toll transmembrane protein during dorsal vessel formation in Drosophila melanogaster
Abstract
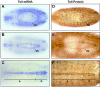
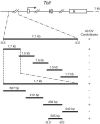
Early heart development in Drosophila and vertebrates involves the specification of cardiac precursor cells within paired progenitor fields, followed by their movement into a linear heart tube structure. The latter process requires coordinated cell interactions, migration, and differentiation as the primitive heart develops toward status as a functional organ. In the Drosophila embryo, cardioblasts emerge from bilateral dorsal mesoderm primordia, followed by alignment as rows of cells that meet at the midline and morph into a dorsal vessel. Genes that function in coordinating cardioblast organization, migration, and assembly are integral to heart development, and their encoded proteins need to be understood as to their roles in this vital morphogenetic process. Here we prove the Toll transmembrane protein is expressed in a secondary phase of heart formation, at lateral cardioblast surfaces as they align, migrate to the midline, and form the linear tube. The Toll dorsal vessel enhancer has been characterized, with its activity controlled by Dorsocross and Tinman transcription factors. Consistent with the observed protein expression pattern, phenotype analyses demonstrate Toll function is essential for normal dorsal vessel formation. Such findings implicate Toll as a critical cell adhesion molecule in the alignment and migration of cardioblasts during dorsal vessel morphogenesis.
Figures





References
-
- Alvarez, A. D., W. Shi, B. A. Wilson, and J. B. Skeath. 2003. pannier and pointedP2 act sequentially to regulate Drosophila heart development. Development 130:3015-3026. - PubMed
-
- Anderson, K. V., G. Jurgens, and C. Nusslein-Volhard. 1985. Establishment of dorsal-ventral polarity in the Drosophila embryo: Genetic studies on the role of the Toll gene product. Cell 42:779-789. - PubMed
-
- Azpiazu, N., and M. Frasch. 1993. tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 7:1325-1340. - PubMed
-
- Barolo, S., L. A. Carver, and J. W. Posakony. 2000. GFP and β-Galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. BioTechniques 29:726-732. - PubMed
-
- Bodmer, R. 1993. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development 118:719-729. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
