The 2 microm plasmid causes cell death in Saccharomyces cerevisiae with a mutation in Ulp1 protease
- PMID: 15870298
- PMCID: PMC1087720
- DOI: 10.1128/MCB.25.10.4299-4310.2005
The 2 microm plasmid causes cell death in Saccharomyces cerevisiae with a mutation in Ulp1 protease
Abstract
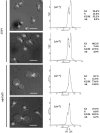
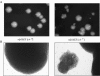
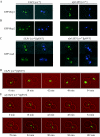
The 2 microm circle plasmid confers no phenotype in wild-type Saccharomyces cerevisiae but in a nib1 mutant, an elevated plasmid copy number is associated with cell death. Complementation was used to identify nib1 as a mutant allele of the ULP1 gene that encodes a protease required for removal of a ubiquitin-like protein, Smt3/SUMO, from protein substrates. The nib1 mutation replaces conserved tryptophan 490 with leucine in the protease domain of Ulp1. Complete deletion of ULP1 is lethal, even in a strain that lacks the 2 microm circle. Partial deletion of ULP1, like the nib1 mutation, results in clonal variations in plasmid copy number. In addition, a subset of these mutant cells produces lineages in which all cells have reduced proliferative capacity, and this phenotype is dependent upon the presence of the 2 microm circle. Segregation of the 2 microm circle requires two plasmid-encoded proteins, Rep1 and Rep2, which were found to colocalize with Ulp1 protein in the nucleus and interact with Smt3 in a two-hybrid assay. These associations and the observation of missegregation of a fluorescently tagged 2 microm circle reporter plasmid in a subset of ulp1 mutant cells suggest that Smt3 modification plays a role in both plasmid copy number control and segregation.
Figures




References
-
- Bachant, J., A. Alcasabas, Y. Blat, N. Kleckner, and S. J. Elledge. 2002. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of topisomerase II. Mol. Cell 9:1169-1182. - PubMed
-
- Bartel, P., C. T. Chien, R. Sternglanz, and S. Fields. 1993. Using the two-hybrid system to detect protein-protein interactions., p. 153-179. In D. A. Hartley (ed.), Cellular interactions in development: a practical approach. IRL Press, Oxford, United Kingdom.
-
- Blat, Y., and N. Kleckner. 1999. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98:249-259. - PubMed
-
- Boddy, M. N., K. Howe, L. D. Etkin, E. Solomon, and P. S. Freemont. 1996. Pic-1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene 13:971-982. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
