Identification of unique type II polyketide synthase genes in soil
- PMID: 15870305
- PMCID: PMC1087561
- DOI: 10.1128/AEM.71.5.2232-2238.2005
Identification of unique type II polyketide synthase genes in soil
Abstract
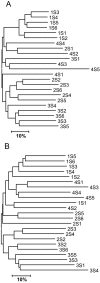
Many bacteria, particularly actinomycetes, are known to produce secondary metabolites synthesized by polyketide synthases (PKS). Bacterial polyketides are a particularly rich source of bioactive molecules, many of which are of potential pharmaceutical relevance. To directly access PKS gene diversity from soil, we developed degenerate PCR primers for actinomycete type II KS(alpha) (ketosynthase) genes. Twenty-one soil samples were collected from diverse sources in New Jersey, and their bacterial communities were compared by terminal restriction fragment length polymorphism (TRFLP) analysis of PCR products generated using bacterial 16S rRNA gene primers (27F and 1525R) as well as an actinomycete-specific forward primer. The distribution of actinomycetes was highly variable but correlated with the overall bacterial species composition as determined by TRFLP. Two samples were identified to contain a particularly rich and unique actinomycete community based on their TRFLP patterns. The same samples also contained the greatest diversity of KS(alpha) genes as determined by TRFLP analysis of KS(alpha) PCR products. KS(alpha) PCR products from these and three additional samples with interesting TRFLP pattern were cloned, and seven novel clades of KS(alpha) genes were identified. Greatest sequence diversity was observed in a sample containing a moderate number of peaks in its KS(alpha) TRFLP. The nucleotide sequences were between 74 and 81% identical to known sequences in GenBank. One cluster of sequences was most similar to the KS(alpha) involved in ardacin (glycopeptide antibiotic) production by Kibdelosporangium aridum. The remaining sequences showed greatest similarity to the KS(alpha) genes in pathways producing the angucycline-derived antibiotics simocyclinone, pradimicin, and jasomycin.
Figures


References
-
- Allsop, A., and R. Illingworth. 2002. The impact of genomics and related technologies on the search for new antibiotics. J. Appl. Microbiol. 92:7-12. - PubMed
-
- Borders, D. B. 1999. Isolation and identification of small molecules, p. 257-265. In A. L. Demain, and J. E. Davies (ed.), Manual of industrial microbiology and biotechnology, 2nd ed. ASM Press, Washington, D.C.
-
- Doull, J. L., A. K. Singh, M. Hoare, and S. W. Ayer. 1994. Conditions for the production of jadomycin B by Streptomyces venezuelae ISP5230: effects of heat shock, ethanol treatment and phage infection. J. Indust. Microbiol. 13:120-125. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

