Heterologous expression of novobiocin and clorobiocin biosynthetic gene clusters
- PMID: 15870333
- PMCID: PMC1087579
- DOI: 10.1128/AEM.71.5.2452-2459.2005
Heterologous expression of novobiocin and clorobiocin biosynthetic gene clusters
Abstract
A method was developed for the heterologous expression of biosynthetic gene clusters in different Streptomyces strains and for the modification of these clusters by single or multiple gene replacements or gene deletions with unprecedented speed and versatility. Lambda-Red-mediated homologous recombination was used for genetic modification of the gene clusters, and the attachment site and integrase of phage phiC31 were employed for the integration of these clusters into the heterologous hosts. This method was used to express the gene clusters of the aminocoumarin antibiotics novobiocin and clorobiocin in the well-studied strains Streptomyces coelicolor and Streptomyces lividans, which, in contrast to the natural producers, can be easily genetically manipulated. S. coelicolor M512 derivatives produced the respective antibiotic in yields comparable to those of natural producer strains, whereas S. lividans TK24 derivatives were at least five times less productive. This method could also be used to carry out functional investigations. Shortening of the cosmids' inserts showed which genes are essential for antibiotic production.
Figures
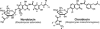




References
-
- Bentley, S. D., K. F. Chater, A. M. Cerdeño-Tárraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. - PubMed
-
- Chen, H., and C. T. Walsh. 2001. Coumarin formation in novobiocin biosynthesis: β-hydroxylation of the aminoacyl enzyme tyrosyl-S-NovH by a cytochrome P450 NovI. Chem. Biol. 8:301-312. - PubMed
-
- Eustáquio, A. S., B. Gust, T. Luft, S.-M. Li, K. F. Chater, and L. Heide. 2003. Clorobiocin biosynthesis in Streptomyces: identification of the halogenase and generation of structural analogs. Chem. Biol. 10:279-288. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

