Polychlorinated biphenyl rhizoremediation by Pseudomonas fluorescens F113 derivatives, using a Sinorhizobium meliloti nod system to drive bph gene expression
- PMID: 15870360
- PMCID: PMC1087564
- DOI: 10.1128/AEM.71.5.2687-2694.2005
Polychlorinated biphenyl rhizoremediation by Pseudomonas fluorescens F113 derivatives, using a Sinorhizobium meliloti nod system to drive bph gene expression
Abstract
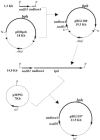
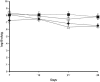
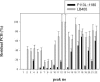
Rhizoremediation of organic chemicals requires high-level expression of biodegradation genes in bacterial strains that are excellent rhizosphere colonizers. Pseudomonas fluorescens F113 is a biocontrol strain that was shown to be an excellent colonizer of numerous plant rhizospheres, including alfalfa. Although a derivative of F113 expressing polychlorinated biphenyl (PCB) biodegradation genes (F113pcb) has been reported previously, this strain shows a low level of bph gene expression, limiting its rhizoremediation potential. Here, a high-level expression system was designed from rhizobial nod gene regulatory relays. Nod promoters were tested in strain F113 by using beta-galactosidase transcriptional fusions. This analysis showed that nodbox 4 from Sinorhizobium meliloti has a high level of expression in F113 that is dependent on an intact nodD1 gene. A transcriptional fusion of a nodbox cassette containing the nodD1 gene and nodbox 4 fused to a gfp gene was expressed in the alfalfa rhizosphere. The bph operon from Burkholderia sp. strain LB400 was cloned under the control of the nodbox cassette and was inserted as a single copy into the genome of F113, generating strain F113L::1180. This new genetically modified strain has a high level of BphC activity and grows on biphenyl as a sole carbon and energy source at a growth rate that is more than three times higher than that of F113pcb. Degradation of PCBs 3, 4, 5, 17, and 25 was also much faster in F113L::1180 than in F113pcb. Finally, the modified strain cometabolized PCB congeners present in Delor103 better than strain LB400, the donor of the bph genes used.
Figures

Similar articles
-
Degradation of PCB congeners by bacterial strains.Appl Microbiol Biotechnol. 2007 Nov;77(2):469-81. doi: 10.1007/s00253-007-1175-6. Epub 2007 Sep 21. Appl Microbiol Biotechnol. 2007. PMID: 17885752
-
Construction of a rhizosphere pseudomonad with potential to degrade polychlorinated biphenyls and detection of bph gene expression in the rhizosphere.Appl Environ Microbiol. 1995 May;61(5):1946-52. doi: 10.1128/aem.61.5.1946-1952.1995. Appl Environ Microbiol. 1995. PMID: 7646029 Free PMC article.
-
Alginate beads as a storage, delivery and containment system for genetically modified PCB degrader and PCB biosensor derivatives of Pseudomonas fluorescens F113.J Appl Microbiol. 2011 May;110(5):1351-8. doi: 10.1111/j.1365-2672.2011.04993.x. Epub 2011 Apr 4. J Appl Microbiol. 2011. PMID: 21395945
-
Whole-cell fluorescent biosensors for bioavailability and biodegradation of polychlorinated biphenyls.Sensors (Basel). 2010;10(2):1377-98. doi: 10.3390/s100201377. Epub 2010 Feb 21. Sensors (Basel). 2010. PMID: 22205873 Free PMC article. Review.
-
Molecular genetics and evolutionary relationship of PCB-degrading bacteria.Biodegradation. 1994 Dec;5(3-4):289-300. doi: 10.1007/BF00696466. Biodegradation. 1994. PMID: 7765839 Review.
Cited by
-
Genome sequence of the biocontrol strain Pseudomonas fluorescens F113.J Bacteriol. 2012 Mar;194(5):1273-4. doi: 10.1128/JB.06601-11. J Bacteriol. 2012. PMID: 22328765 Free PMC article.
-
Bacterial metabolism of polycyclic aromatic hydrocarbons: strategies for bioremediation.Indian J Microbiol. 2008 Mar;48(1):95-113. doi: 10.1007/s12088-008-0010-9. Epub 2008 May 1. Indian J Microbiol. 2008. PMID: 23100704 Free PMC article.
-
Polychlorinated Biphenyl Transformation, Peroxidase and Oxidase Activities of Fungi and Bacteria Isolated from a Historically Contaminated Site.Microorganisms. 2023 Jul 26;11(8):1887. doi: 10.3390/microorganisms11081887. Microorganisms. 2023. PMID: 37630447 Free PMC article.
-
Computational prediction of the Crc regulon identifies genus-wide and species-specific targets of catabolite repression control in Pseudomonas bacteria.BMC Microbiol. 2010 Nov 25;10:300. doi: 10.1186/1471-2180-10-300. BMC Microbiol. 2010. PMID: 21108798 Free PMC article.
-
Plant-associated bacterial degradation of toxic organic compounds in soil.Int J Environ Res Public Health. 2009 Aug;6(8):2226-47. doi: 10.3390/ijerph6082226. Epub 2009 Aug 12. Int J Environ Res Public Health. 2009. PMID: 19742157 Free PMC article. Review.
References
-
- Baev, N., M. Amar, R. Defez, and M. Iaccarino. 1992. The expression of the nodD and nodABC genes of Rhizobium leguminosarum is not regulated in response to combined nitrogen. FEMS Microbiol. Lett. 97:205-208.
-
- Baev, N., G. Endre, G. Petrovics, Z. Banfalvi, and A. Kondorosi. 1991. 6 nodulation genes of nod box locus-4 in Rhizobium meliloti are involved in nodulation signal production—nodM codes for d-glucosamine synthetase. Mol. Gen. Genet. 228:113-124. - PubMed
-
- Ballschmiter, K., and M. Zell. 1980. Analysis of polychlorinated biphenyls (PCB) by glass-capillary gas chromatography—composition of technical Aroclor-PCB and Clophen-PCB mixtures. Fresenius Z. Anal. Chem. 302:20-31.
-
- Barea, J. M., G. Andrade, V. Bianciotto, D. Dowling, S. Lohrke, P. Bonfante, F. O'Gara, and C. Azcon-Aguilar. 1998. Impact on arbuscular mycorrhiza formation of Pseudomonas strains used as inoculants for biocontrol of soil-borne fungal plant pathogens. Appl. Environ. Microbiol. 64:2304-2307. - PMC - PubMed
-
- Barriault, D., and M. Sylvestre. 1993. Factors affecting PCB degradation by an implanted bacterial strain in soil microcosms. Can. J. Microbiol. 39:594-602. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

