Adeno-associated virus: from defective virus to effective vector
- PMID: 15877812
- PMCID: PMC1131931
- DOI: 10.1186/1743-422X-2-43
Adeno-associated virus: from defective virus to effective vector
Abstract
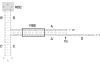
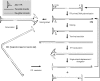
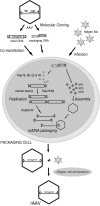
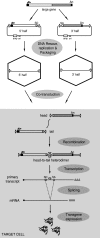
The initial discovery of adeno-associated virus (AAV) mixed with adenovirus particles was not a fortuitous one but rather an expression of AAV biology. Indeed, as it came to be known, in addition to the unavoidable host cell, AAV typically needs a so-called helper virus such as adenovirus to replicate. Since the AAV life cycle revolves around another unrelated virus it was dubbed a satellite virus. However, the structural simplicity plus the defective and non-pathogenic character of this satellite virus caused recombinant forms to acquire centre-stage prominence in the current constellation of vectors for human gene therapy. In the present review, issues related to the development of recombinant AAV (rAAV) vectors, from the general principle to production methods, tropism modifications and other emerging technologies are discussed. In addition, the accumulating knowledge regarding the mechanisms of rAAV genome transduction and persistence is reviewed. The topics on rAAV vectorology are supplemented with information on the parental virus biology with an emphasis on aspects that directly impact on vector design and performance such as genome replication, genetic structure, and host cell entry.
Figures

References
-
- Atchison RW, Castro BC, Hammon WM. Adenovirus-associated defective virus particles. Science. 1965;149:754–756. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

